Ty1/Copia
Introduction
The Ty1/Copia depicts one of the most important and representative families of LTR retroelements in eukaryotes (see also, Eickbush and Malik 2002). The abundant representation of Ty1/Copia LTR retroelements in the genomes of plants, fungi, animals, algae and several protists suggests that the ancestors of this family probably co-existed with the ancestors of Ty3/Gypsy LTR retroelements before the split between plants and unikonts (see, Llorens et al. 2009 and references therein). Although the LTR retrotransposon form is what predominates in the Ty1/Copia family it is now known that several Ty1/Copia elements described in plants (those belonging to Sire clade) are carriers of a third ORF env which confer them the status of potential retrovirus (Havecker et al. 2005). To date, there is no evidence about retrovirus-like Ty1/Copia representatives in fungal and metazoan organisms.
Genomic structure:
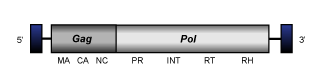
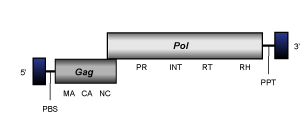
Ty1/Copia LTR retroelements show identical functions and similar genome organization than other LTR retroelements. That usually is:
- A 5' long terminal repeat (LTR) of 100-1300 nt
- A non-coding region of variable size that corresponds to the first portion of the retrotranscribed genome.
- A Primer Binding Site (PBS) of 18 nt, complementary to a specific zone of the 3' end of a tRNA normally provided by the host cell in order to start the retrotranscription.
- Open Reading Frames (ORFs) for gag, pol, (and env in retroviruses) genes.
- A small region of ~10 A/G "Polypurine Tract" (PPT), responsible for starting the synthesis of the proviral DNA strand.
- A 3' long terminal repeat (LTR) of 100-1300 nt
However, Ty1/Copia elements differ from other other LTR retroelements in the position of integrase (INT) domain within pol polyprotein. While Bel/Pao, Retroviridae and almost but not all Ty3/Gypsy LTR retroelements show the INT at the C-terminus of pol (after RNase H), Ty1/Copia elements present INT N-terminal to the reverse transcriptase (after the protease domain) (for a review, see Eickbush and Jamburuthugoda 2008; see also LTR retroelements).

Evolutionary history and in-progress taxonomy
The classification of Ty1/Copia family is work in progress. According to the International Committee on the Taxonomy of Viruses (ICTV) the Ty1/Copia family is called Pseudoviridae and was originary divided into three genera namely, Pseudovirus (type species Saccharomyces cerevisiae Ty1 element, SceTy1V), Hemivirus (type species Drosophila melanogaster Copia element, DmeCV) and Sirevirus (type species Glycine max SIRE-1 virus, GmaSIRE-1V) (Boeke et al. 2005). Pseudoviruses and hemiviruses are normally distinguished by the primer used for reverse transcription (a full tRNA or a half tRNA, respectively) while sireviruses derive from plant hosts and make up a distinct cluster according to their RT amino-acid sequences (Havecker et al. 2004).
The ICTV classification is important for understanding the original Ty1/Copia family but it is not conclusive to date for managing the currently available diversity of this family. According to (Llorens et al 2009), an update of the phylogeny of Ty1/Copia LTR retroelements based on pol reveals two major branches 1 and 2. Branch 1 collects Pseudovirus genus (see Havecker et al. 2004) together with a clade called GalEA (Terrat et al. 2008), found in marine bilaterians, and all CoDi-like LTR retrotransposons found in diatom genomes (Bowler et al. 2008; Maumus et al. 2009). According to Llorens et al. 2009, the wide variety of CoDi-like elements splits into four clades that we simply name I, II (or A and B), C and D. One of the most exciting aspects of CoDi-like elements is that the different elements belonging to clade A encode INTs carrying a putative chromodomain at their C-terminus (Llorens et al. 2009). The second Ty1/Copia branch encompasses the remaining lineages of LTR retroelements, including the previously described Copia-like hemiviruses and sireviruses. A description follows in the table and discussion below.
| Branch | Host Phyla | Genus | Clade | Env | Chromodomain |
|---|---|---|---|---|---|
| Branch 1 | Fungi | Pseudovirus (Pseudoviridae) | Ty (Pseudovirus) | no | |
| Branch 1 | Diatoms (Heterokontophyta) | CoDi-I or CoDi-A | no | ||
| Branch 1 | Diatoms (Heterokontophyta) | CoDi-II or CoDi-B | no | ||
| Branch 1 | Diatoms (Heterokontophyta) | CoDi-C | no | ||
| Branch 1 | Diatoms (Heterokontophyta) | CoDi-D | no | ||
| Branch 1 | Marine Arthropoda | GalEA | no | ||
| Branch 2 | Fungi | p-Cretro | no | ||
| Branch 2 | Land plants (Viridiplantae) | Sirevirus (Pseudoviridae) | Sire | yes | |
| Branch 2 | Land plants (Viridiplantae) | Sirevirus (Pseudoviridae) | Oryco | no | |
| Branch 2 | Land plants (Viridiplantae) | Retrofit | no | ||
| Branch 2 | Land plants (Viridiplantae) | Tork | no | ||
| Branch 2 | Green algae (Viridiplantae) | Osser | no | ||
| Branch 2 | Red algae (Rhodophyta) | PyRE1G1 | no | ||
| Branch 2 | Cnidaria (Metazoa) | Hydra | no | ||
| Branch 2 | Arthropoda | Hemivirus (Pseudoviridae) | Copia | no | |
| Branch 2 | Arthropoda | 1731 | no | ||
| Branch 2 | Arthropoda | Tricopia | no | ||
| Branch 2 | Arthropoda | Mtanga | no | ||
| Branch 2 | Arthropoda | Humnum | no |
Branch 1
The first branch, which we call Branch 1, encompasses the original Pseudovirus genus together with a GalEA clade called, a lineage specifically found in marine bilaterians, and all CoDi-like LTR retrotransposons described in diatoms. The variety of CoDi-like elements splits into four clades that we simply name A, B, (or I and II), C and D (Llorens et al. 2009).
Ty (Pseudovirus)
Pseudovirus elements are Ty1/Copia LTR retrotransposons normally found in fungi (Havecker et al. 2004). The members of this clade usually have a genome of 5.6-6.8 Kb in size, which show an internal gag-pol region flanked by LTRs of 0.3-0.4 Kb and showing the typical Primer Bindind Site (PBS) and one or two Polypurine Tracts (PPT) downstream and upstream to the 5´and 3´LTRs, respectively (Mattews et al. 1997; Friant et al. 1996; Heyman et al. 2003; Neuvéglise et al. 2002). Among Ty retrotransposons only Ty4 elements contain the gag-associated RNA-binding motif (CCHC). This motif has not been identified in the gags of the other members belonging to this clade (Peterson-Burch and Voytas 2002; Neuvéglise et al. 2002).
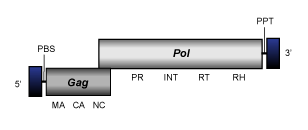
CoDi-I
The elements belonging to clade CoDi-I (Maumus et al. 2009) also called CoDi-A (Llorens et al. 2009) have been described in the genomes of the marine pennate diatom Phaeodactylum tricornutum and the marine algae phytoplankton Thalassiosira pseudonana (Maumus et al. 2009; Bowler et al. 2008). The genome of these Ty1/Copia-like LTR retroelements is about of 5.9-7.4 Kb in size, including LTRs of 0.15-0.69 Kb long that flank a single long ORF. The ORF contains the gag and pol typical Ty1/Copia domains structure (gag, protease, integrase, reverse transcriptase and ribonuclease H). The conserved integrase motif has not been clearly identified in the genome of these elements, a well as the gag-associated zinc finger motif. Interestingly, CoDi-I represents the first described set of Ty1/Copia retroelements carrying a putative chromodomain at the C-terminus of their pol-INT domain (Llorens et al. 2009) similarly to Ty3/Gypsy chromoviruses (Marin and Llorens 2000), which use this feature for chromatin integration (Gao et al. 2008).
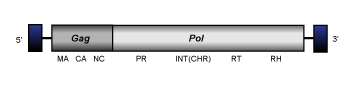
CoDi-II
CoDi-II Ty1/Copia-like LTR retroelements (Maumus et al. 2009), also called CoDi-B (Llorens et al. 2009), have been described in the genomes of the marine pennate diatom Phaeodactylum tricornutum and the marine algae phytoplankton Thalassiosira pseudonana (Maumus et al. 2009; Bowler et al. 2008). These display a genome of about 5.2-6.1 Kb in size, including LTRs of 0.16-0.30 Kb long that bound one or two overlapping ORFs. The translated regions contain both gag and pol Ty1/Copia-like polyproteins domains (gag, protease, integrase, reverse transcriptase and ribonuclease H). The conserved integrase motif has not been clearly identified in the genome of CoDi-II retroelements.
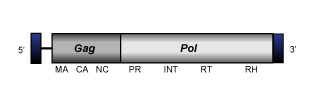
CoDi-C
CoDi-C clade is the term used by Llorens et al. (2009) to differentiate the two clades constituted by the CoDi-6-like elements described by Maumus et al. (2009) in the genomes of the marine pennate diatom Phaeodactylum tricornutum and the marine algae phytoplankton Thalassiosira pseudonana (see also Bowler et al. 2008). The genome of CoDi-C elements, of about of 6.6-8 Kb in size, includes 0.27-0.49 Kb LTRs long flanking one or two overlapping ORFs that show both gag and pol Ty1/Copia-like polyproteins domains (gag, protease, integrase, reverse transcriptase and ribonuclease H). CoDi-6-like elements are the most divergent CoDi-like, their position in the Ty1/Copia phylogeny is still under study as depending on the protein domain evaluated they may fall together with other CoDi-like clades within Branch 1 or close to other Ty1/Copia elements of protostomes (for more details, see the collection of trees provided at GyDB, or (Llorens et al. 2009; Maumus et al. 2009).
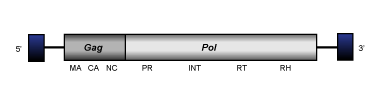
CoDi-D
The term CoDi-D (Llorens et al. 2009) is used to differentiate the two clades constituted by the CoDi-6-like elements described by Maumus et al. (2009) in the genomes of the marine pennate diatom Phaeodactylum tricornutum and the marine algae phytoplankton Thalassiosira pseudonana (see also Bowler et al. 2008). CoDi-D retroelements display a genome of about 5.1-5.3 Kb in size including LTRs of 0.16-0.41 Kb long. The LTRs flanked internal region encodes for a single long ORF that contains both gag and pol Ty1/Copia-like polyproteins domains (gag, protease, integrase, reverse transcriptase and ribonuclease H). In the gag-nucleocapsid domain of CoDi-D elements have been identified two zinc finger motifs. CoDi-6-like elements are the most divergent CoDi-like, their position in the Ty1/Copia phylogeny is still under study as depending on the protein domain evaluated they may fall together with other CoDi-like clades within Branch 1 or close to other Ty1/Copia elements of protostomes (for more details, see the collection of trees provided at GyDB, or (Llorens et al. 2009; Maumus et al. 2009).
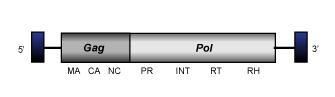
GalEA
This is a lineage of LTR retrotransposons, which seems to be restricted to aquatic species (they have been identified in crustaceans, fishes and urochordates). Its name derives from Galatheids Eumunida annulosa retrotransposons. The distinct members of this clade usually show an internal region of 3.9-4.4 kb in size delimited by LTRs of 0.12 – 0.32 kb, that includes a single large ORF containing gag and pol regions (Terrat et al. 2008).
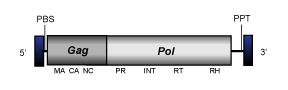
Branch 2
The second Ty1/Copia branch encompasses the remaining lineages of LTR retrotransposons and potential retroviruses, including Copia-like hemiviruses and sireviruses.
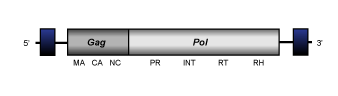
pCretro
The elements belonging to this clade have been described in the genome of lignin-degrading basidiomycete Phanerochaete chrysosporium (Larrondo et al. 2007). The full-lenght consensus of known to date pCretro elements is about of 5 kb in size, contains 5' and 3' LTRs 0.36-0.4 kb long, and shows an internal region that codes for the gag and pol polyproteins characteristic of Ty1/Copia LTR retrotransposons.
Sireviruses
These (Havecker et al. 2004; Havecker et al. 2005) constitute a cluster of LTR retrotransposons and retroviruses described in plants and can be divided in two phylogenetically related lineages "Sire" and "Oryco" (according to Llorens et al. 2009). These two differ in that the elements belonging to Sire clade usually (but not always) code for an additional putative env gene and are considered as potential retroviruses, while Oryco-like elements are small LTR retrotransposons.
Sire
Sire elements are large LTR retroelements showing genomes of 9.3-9.8 kb in size flanked by LTRs of 0.5-1.2kb. Additionally to the typical gag and pol ORFs Sire-like elements show a third env-like ORF downstream to RNase H domain (and are thus considered potential retroviruses, Havecker et al. 2004; Havecker et al. 2005) and encode for a significantly large gag polyprotein, which may has two or three zinc finger arrays at its C-terminus (Havecker et al. 2005). The most representative example of Sire-like element encoding for a putative ENV is SIRE-1, an element originally identified in the soybean Glycine max (Laten et al. 1998; Laten 1999). SIRE1-like elements are widespread among plant species, both in monocots (rice, maize, sorghum) and dicots (Arabidopsis, lotus, medicago, citrus) (Havecker et al. 2005).
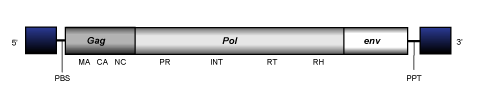
Oryco
These LTR retrotransposons have been found in the genome of some plant species: Vitis vinifera, Arabidopsis thaliana, Popolus tricocarpa and Oryza sativa (Llorens et al. 2009). Their small genome is about of 4.2-4.9 Kb in size included LTRs of 0.16-0.44 Kb long. The internal coding region displays the gag-pol domain order of Ty1/Copia retrotransposons. No env-gene has been detected in their genome.
Retrofit
The plant LTR retrotransposons that constitute Retrofit clade have a genome of about 4.7-4.9 Kb, included small LTRs of 0.12-0.3 kb long, encoding for both gag-pol polyprotein which domain organization is the tipical of Ty1/Copia LTR retrotransposons (Pastuglia et al. 1997; Song et al. 1997; White et al. 1994; Llorens et al. 2009; Piegu et al. 2006).
Tork
Tork clade belong LTR retrotransposons described in the genome of different plant species: Zea mays, Solanum lycopersicum, Vitis vinifera, Nicotiana tabacum and Vigna radiata (SanMiguel et al. 1996; Llorens et al. 2009; Marillonnet and Wessler 1998; Grandbastien et al. 1989; Hirochika et al. 1996; Xiao et al. 2007). The members of this clade show very different size in both LTRs and coding regions being respectively of 0.12-1.2 Kb and 4.1-6.7 Kb long.
Osser
Osser is the first complete Ty1/copia-like retrotransposon described in the colonial green alga Volvox carteri. Its genome is 4.875 bp in size and bordered by two identical LTRs of 197 bp in length. Osser internal region is characterized by a large central domain with the typical gag-pol organization of retrotransposons belonging to the Tyl/copia family (Lindauer et al. 1993) (for more details see the file of Osser element).
PyRE1G1
PyRE1G1 is a Ty1/copia-like retrotransposon constituting a single clade that was characterized in the genome of the red alga Porphyra yezoensis. Its genome is 4807 bp in size including two LTRs of 204 bp. The internal region of this element presents an ORF of 1401 residues that codes for a single gag-pol polyprotein (Peddigari et al. 2008) (for more details see the file of PyRE1G1 element).
Hydra
Hydra clade collect at least two elements - Hydra1-1 and Hydra 1-2 - both described in the genome of the freshwater animal Hydra magnipapillata and in that of the zebra fish Danio rerio (Llorens et al. 2009). The genome of these elements is about of 4.1-4.3 Kb and reveals a single long ORF that, although is broken by stop codons, contains the typical Ty1/Copia gag-pol domains structure. LTRs of 163 bp in size have been described for Hydra1-1, while information about the LTRs of Hydra1-2 is not yet available.
Copia
The elements of Copia clade constitute a clade of LTR retrotransposons described in Insecta (Peterson-Burch and Voytas 2002; Ohbayashi et al. 1998; Yoshioka et al. 1992). Their genome, about of 4.7-5.2Kb in size, includes LTRs of 0.18-0.27 kb bounding an internal region constituted by one or two overlapping ORFs which domain organization is the typical of Ty1/Copia retroelements.
1731
1731 clade is a lineage of diptera LTR retrotransposons, which usually are 4.5-4.6 kb in size and show an internal gag-pol coding region flanked by LTRs of 0.2-0.3 kb long (Fourcade-Peronnet et al. 1988).
Tricopia
Tricopia represents a small clade of LTR retrotransposons described in the genome of the red flour beetle Tribolium castaneum (Llorens et al. 2009) and shows the typical Ty1/Copia genome structure: a translated region containing the gag-pol domains flanked by 0.25-0.26-bp LTRs. The Tricopia full-length genome is 4541 bp long (for more details see the file of Tricopia element).
Mtanga
Mtanga represents a clade of Ty1/Copia LTR retrotransposons described from the African malaria vector, Anopheles gambiae. As they are specific to the Y chromosome of this mosquito they are designated as mtanga-Y elements. The genome of these elements normally is 4.284 bp and reveals two overlapping gag and pol genes bounded by LTRs of 119 bp in size (Rohr et al. 2002) (for more details see the file of Mtanga element).
Humnum
Humnum is a Ty1/copia-like retrotransposon found in the genome of the Lepidoptera Heliconius numata (Llorens et al. 2009). Humnum genome, of about of 4.4 Kb in size, includes 139-bp LTRs flanking a single long polyprotein. The encoded polyprotein contains both gag and pol Ty1/Copia associated domains (for more details see the file of Humnum element).
