Intro
GyDB project
The GyDB project pursues the fascinating goal of analyzing and classifying the non-redundant diversity of mobile genetic elements in the context of the tree of life and based on their evolutionary profiles. The emergence and subsequent role played by viruses and mobile genetic elements in the history of life is an unclear yet exciting object of investigation, where researchers try to discern relevant aspects of the molecular changes responsible for various characteristics in the biological organisms related with horizontal transfer, infection and disease.
The most captivating aspect of our project is that we dedicate a share of our efforts to the interpretation of our analyses, we pay special attention to non-redundant elements displaying a certain degree of distance and how they can be collectively aligned or related, in terms of protein domain architecture with other lineages and elements. This is an effort worth making, as we are able to infer the evolutionary perspectives of the elements we classify based on the common internal region. The GyDB project is thus a highly informative database established within an evolutionary context of classification. We think this strategy will allow the GyDB to enable exciting insights leading to a better understanding of the taxonomy and evolutionary history of the elements we classify. Hence, although you may detect imperfections and lacks (new evidence concerning mobile genetic elements is continuous), this is however, what makes this project exciting; one research delivers one conclusion that drives you to another goal, the principal aim which with we have built this database.
Due to their impressive molecular diversity, the GyDB is a long-term project that has been arranged in a database in continuous progress that must be achieved in stages. Taking this into primary consideration this second release (GyDB 2.0) has been powered by a wiki content management system to let other researchers participate in the project. The current database scope is LTR retroelements and their relatives. On the basis of this scope, the first release focused on the evolutionary relationships of Ty3/Gypsy and Retroviridae LTR retroelements, and clan AA of Aspartic Peptidases. GyDB 2.0 is an update based on the analysis of Ty3/Gypsy, Retroviridae, Ty1/Copia and Bel/Pao LTR retroelements and the Caulimoviridae pararetroviruses of plants.
Mobile Genetic Elements
In a broad sense, Mobile Genetic Elements (MGEs) can be considered as segments of DNA somehow involved in the movement of DNA within genomes (intracellular mobility) and between cells (intercellular mobility) (Frost et al. 2005). Such definition would include genomic islands, bacteriophages and plasmids along with transposable elements (TEs). The latter are initially the elements we are going to focus on at GyDB considering MGEs as self-contained genomic units capable of proliferating within their host genomes.
Since the existence of mobile DNA was first suggested by McClintock (McClintock 1948) MGEs have been important object of study in multiple areas of research (Kazazian 2004). Nearly all fit into two major categories of TEs classified by their mechanisms:
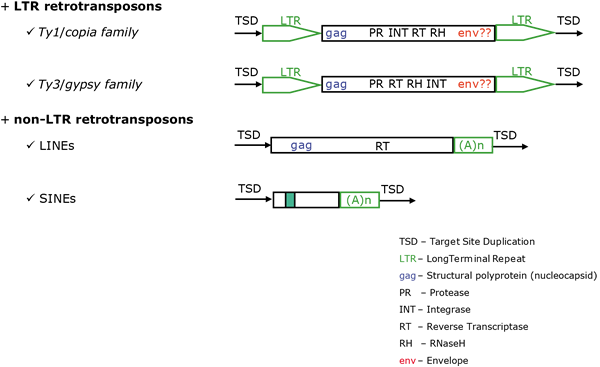
- Class I are retroelements (Temin 1989) that mediate their transposition life cycle through an RNA-DNA reverse transcription process. In collective, all reverse transcriptase (RT) dependent retrotransposons and retroviruses (detailed description concerning this group will be found at Retroelements) .

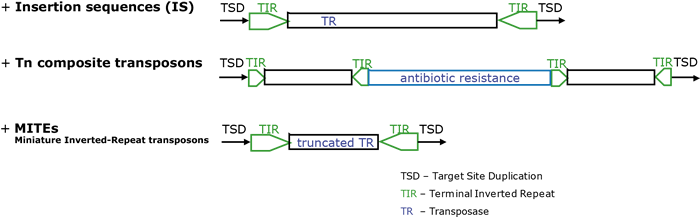
- Class II are DNA-based transposons that move directly from one position to another in host genomes (McClintock 1948; Mizuuchi 1992; Craig et al. 2002).

Transposable Elements are ubiquitous in nature, quite abundant and irregularly distributed within genomes. They can act as mutagenic agents, generate DNA rearrangements, influence gene expression and, in short, play an important role in the evolution of the host genome. Identification, characterization and analysis of TEs and their dynamics are important for a better understanding of the structure and evolution of both, genomes and mobile DNA themselves (Holmes 2002; Kidwell & Lisch 1997). However, scientific interest on TEs lies not only in derivative evolutive studies (as they are considered fossil markers), but also in its role in host genomes, its usefulness as biotechnological tools even for genetic medical applications and other potential future implications (Levin & Moran 2011; Nesmelova & Hackett 2010; Hickman et al. 2005).
