Protease
LTR retroelement-like proteases (PRs, also named here as APs) are proteolytic enzymes that play a key role in the maturation process during which several peptides involved in the life cycle of the retroelement are scissed by this enzyme. LTR retroelement PRs belong to clan AA of aspartic peptidases (Rawlings et al. 2008); they dimerize in their active form and may be encoded as a part of the pol polyprotein, alone or as a part of the gag polyprotein, or in frame with a dUTPase (see dUTPase section). It is well known that the structural PR homodomain is founded in a core ~90-150 residues long wherein the catalytic DTG motif (Pearl and Blundell 1984) is the most prominent feature along with a glycine at the C-terminal end preceded by two hydrophobic residues (Pearl and Taylor 1987).
At the primary structure level the most conserved part (core) of all clan peptidases may be divided in six amino acidic patterns constituting a template we have called "DTG/ILG". We introduce this template in a forthcoming study (see the references below) but may be preliminarily compared as a HMM profile via the HMM server of this database.
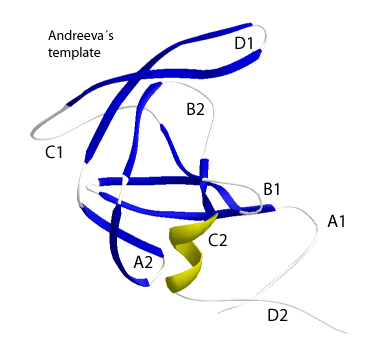
The "DTG/ILG" template is the primary structure phenotype of a structural supersecondary structure, called "Andreeva’s" template (Andreeva 1991) that was previously used to describe pepsins and retropepsin. The "Andreeva’s" template is constituted by the following structural elements: an N-terminal loop (A1), a loop containing the catalytic motif (B1), an α-helix (C1) usually not preserved in retropepsins, a β-hairpin loop (D1), a hairpin loop (A2), a wide loop (B2), an α-helix (C2) towards C-terminal, and a loop (D2), which in empirically characterized retropepsins is substituted by a strand or a helical turn (Wlodawer and Gustchina 2000; Dunn et al. 2002). These elements are responsible of keep both function and three-dimensional (3D) structure in characterized retropepsins and other characterized clan AA peptidases (Wlodawer and Gustchina 2000; Dunn et al. 2002). It has also recently suggested that the structure of the HIV-1 (see the figure below) and other clan AA PRs have a flexibility-assisted mechanism evolutionarily preserved to favor the reactive conformation of the enzyme (Piana, Carloni, and Rothlisberger 2002; Piana, Carloni, and Parrinello 2002; Perryman, Lin, and McCammon 2004).