Clan AA
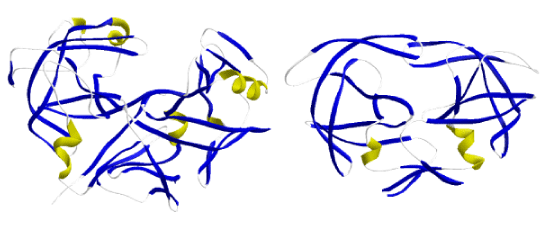
Clan AA of aspartic peptidases (CAPs) is a group of proteolytic enzymes (Rawlings et al. 2008) that use an aspartate dyad and a molecule of water to hydrolyze a peptide bond (Fruton 1976). As shown in the figure below, the different enzymes belonging to clan AA can be divided in two large structural subgroups (Davies 1990; Rawlings et al. 2008), the first comprises all nonviral eukaryotic pepsin monomers structured in two protein domains of similar architecture, the second embraces all the single domain proteases that dimerize in their active form and are usually part of the pol polyprotein coded by LTR retrotransposons and retroviruses. It is usually assumed that the two-domain form of pepsins evolved from the gene duplication of a single ancestor (Tang et al. 1978) related to retroviral-like CAPs (Pearl and Taylor 1987). This is because the similar pseudo symmetry of both dimeric and monomeric CAPs (see Wlodawer and Gustchina 2000).
The HIV-1 CAP and other retropepsins and pepsins have been studied and compared through structure-based alignments (Pearl and Taylor 1987; Weber 1989) and other comparisons (Wlodawer et al. 1989; Rao, Erickson, and Wlodawer 1991;Wlodawer and Gustchina 2000; Dunn et al. 2002; Cascadella et al. 2005; Li et al. 2005; Jaskolski et al. 2006). Upon that, the most prominent phenotype of almost but not all CAPs is a catalytic DT/SG triad (Pearl and Blundell 1984) displayed near to the N-terminus, and a glycine, preceded by two hydrophobic residues (normally isoleucine and leucine) close to the C-terminus of the peptidase core (Pearl and Taylor 1987). This core exhibits a poorly preserved hydrophobic architecture of ~90-150 residues in length (Pearl and Taylor 1987; Weber 1989), which usually follows a structural template rich in β-strands originally introduced by Andreeva (1991) to describe pepsins. The "Andreeva's template" thus defines the peptidase fold in all empirically characterized CAPs (Wlodawer et al. 1989; Rao, Erickson, and Wlodawer 1991; Wlodawer and Gustchina 2000; Dunn et al. 2002; Cascadella et al. 2005; Li et al. 2005; Jaskolski et al. 2006).
An important principle of peptidase classification has been established by MEROPS (Rawlings et al. 2008) (a general database on enzymology), according to which pepsins and LTR retroelement CAPs can be divided in five families: A1, A2, A3, A9 and A11 (according to the table below). Here, pepsins represent the family A1, which splits in 2 subfamilies - A1A and A1B. MEROPS classify the different CAPs encoded by vertebrate retroviruses (Retroviridae) as the family A2 (retropepsins) except those encoded by Spumaretroviruses, which were assigned to the family A9 (spumaretropepsins). MEROPS also classifies a few examples of Ty3/Gypsy CAPs in several sub-families within the family A2 because of their similarity to retropepsins. However, not all Ty3/Gypsy CAPs are similar to retropepsins just as not all the Retroviridae CAPs are retropepsins. The different CAPs encoded by Caulimoviridae and Ty1/Copia LTR retroelements were assigned to the families A3 and A11 respectively. The CAPs encoded by Bel/Pao LTR retroelements have no current family classification (for a more detailed perspective about the aforesaid LTR retroelement groups, see Eickbush and Malik 2002).
| Letter | Family | Domain | Genome |
|---|---|---|---|
| A1 | Pepsins | double domain | non-viral |
| A2 | Retropepsins | homodomain | retroelement-like |
| A3 | Caulimoviral proteases | homodomain | retroelement-like |
| A9 | Spumaretropepsins | homodomain | retroelement-like |
| A11 | Ty1/Copia proteases | homodomain | retroelement-like |
In the post-genomic era, sequencing projects have revealed how the diversity of clan AA (i.e. the potential clan families and subfamilies) clearly exceeds the current classification. In fact, the split between the two structural forms of clan AA (dimeric and monomeric) is probably older than previously supposed as sequencing projects have recently revealed the presence of pepsin representatives in several prokaryotic genomes (see MEROPS Database) and two sets of single-domain nonviral CAPs in prokaryotes (COG3577 and COG5550) and three others in eukaryotes (DDI, NIX1 and SASPases) (Krylov and Koonin 2001; Puente et al. 2003; Bernard et al. 2005; Matsui et al. 2006). Having recognized that, the large diversity of clan AA clearly justifies the development and continuous update of a more detailed classification focussing on the different peptidases encoded by eukaryotic LTR retroelements and related host genes. With this aim, we have created the Clan AA Reference Database (CAARD), an in progress database created to investigate the major consensus and the phylogeny of clan AA to typify the different protein families by sequence logos and HMMs. In this first version, we perform a non-redundant evaluation of all clan AA families, but pay particular attention to Ty3/Gypsy and Retroviridae CAPs according to their differentiation into clades and genera. The database is in continuous progress and we are committed to perform similar investigation based on other LTR retroelement groups.