Reverse Transcriptase
The Reverse Transcriptase (RT) is an enzyme capable of catalyzing the synthesis of DNA from a single strain of RNA or DNA (Baltimore 1970; Temin and Mizutani 1970). This enzyme has been associated with Telomerase Reverse Transcriptases (TERTs) involved in the replication of the ends of eukaryotic organisms’ chromosomes (see review Nakamura and Cech 1998).
The reverse-transcription process is common among a wide range of prokaryotic and eukaryotic mobile genetic elements, and requires a primer of 12-18 bases in length usually provided by the 3´end of a host tRNA (Eickbush 1994). As an evolutionary relationship is commonly assumed to exist among all RT-dependent mobile genetic elements, they have been collectively termed Retroelements (Temin 1989).
At the primary structure level, RTs codified by Ty3/Gypsy and Retroviridae elements expand approximately 350 residues of the pol polyprotein, including an alignable core of approximately 180 aa wherein seven conserved regions can be distinguished (Xiong and Eickbush 1988; 1990).
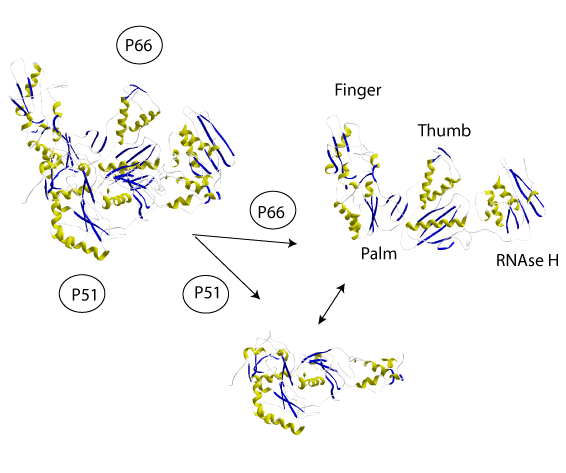
At the three-dimensional (3D) structure level the RT codified by the HIV-1 retrovirus is an asymmetrical heterodimer composed of two subunits of 66 and 51 kDa, p66 and p51 respectively. P66 can be divided into five structural subdomains consisting in the RNaseH domain and four subdomains which, due to their similarity to a human right hand, are referred to as fingers, palm, thumb, and connection (Kohlstaedt et al. 1992). P51 is a p-66´ derivative after proteolytic processing and excision of the RNase H (Di Marzo et al. 1986; Wondrak et al. 1986; Lightfoote et al. 1986).
Although several evidences indicate that RTs encoded by other vertebrate retroviruses also form a heterodimer, the RT may also be functionally active as a monomer (see Misra, Pandey and Pandey 1998).