Movement protein
The movement (MOV) protein typically encoded by Caulimoviridae viruses is involved in the cell-to-cell movement, which is an essential step in plant virus infection. Movement is the process by which diverse groups of plant RNA and DNA viruses spread from initially infected cells to other uninfected cells, up to whole organs within the host organism (for a review, see Heinlein 2009). Indeed, plant viruses use a number of movement pathways inside their hosts (Carrington et al. 1996; Lazarowitz and Beachy 1999). The most common intercellular movement is the "cell-to-cell trafficking" through plasmodesmata (PD), which are plant-specific intercellular cytoplasmic bridges that provide continuity between adjacent cells and tissues (Oparka 2004). The interaction between the virus and the PD is mediated by specific virus-encoded non-cell-autonomous proteins (NCAPs). The firstly identified NCAP was a 30 kDa non-structural protein (30K) encoded by the genome of the single-stranded RNA Tobacco mosaic virus (TMV) the type species of the genus Tobamovirus. This protein has been the first described plant movement protein (for a review, see Heinlein 2009) to be involved in "cell-to-cell trafficking" through PD (Tomenius et al. 1987). Additionally, plant viruses can also move among distinct organs (long-distance transport) via phloem through the vascular system, which is the system used by plants for the transport of assimilates and macromolecules (van Bel 2003a, 2003b). The different between these two mechanisms is that cell-to-cell trafficking is an active intracellular process that requires specific interaction between the virus and PD while the virus long-distance transport is a passive mechanism occurring with the flow of assimilated.
In the case of TMV (the virus species most extensively used as model study), MOV interacts with PD modifying their size exclusion limit (SEL) and permitting the passage of MOV molecules (Waigmann et al. 1994) as well as of a ribonucleoprotein complex (vRNP) composed of anon-encapsidated viral RNAs (vRNAs) and MOVs (Dorokhov et al. 1984; Sambade et al. 2008). Regarding Caulimoviruses, well studied species such as the Cauliflower mosaic virus (CaMV) and the Commelina yellow mottle virus (ComYMV) use a different mechanism to move within the host. Caulimoviral MOVs constitute tubular transporting structures within the PD through which the encapsidated virions pass (Linstead et al. 1988; Cheng et al. 1998; Sánchez-Navarro et al. 2010). Upon this, it has been suggested that some viruses may have the option to use both distinct movement strategies, as observed in TMV (Heinlein et al. 1998) and CaMV (Carrington et al. 1996). Additionally, although the two aforementioned means probably represent the two major strategies for virus cell-to-cell movement, intermediate modes have been also described (Canto and Palukaitis 2005; Kaplan et al. 1998; Sanchez-Navarro and Bol 2001).
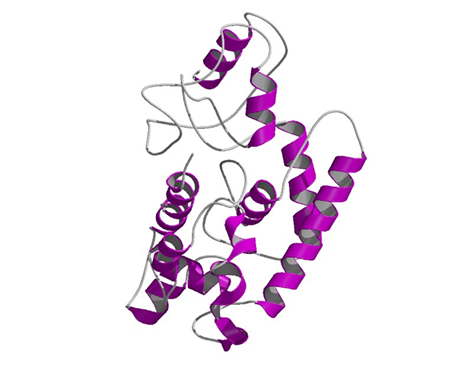
Caulimoviridae can encode the movement protein as a single gene (Linstead et al. 1988; Hasegawa et al. 1989) or as a part of a large polyprotein including retroelement-like domains such as the Gag-coat or pol (Cheng et al. 1998; Richert-Pöggeler and Shepherd 1997; Hull 1996). Sequence comparisons of plant MOV proteins belonging to the principal groups of (+) strand RNA viruses, as well as ssDNA geminiviruses, dsDNA pararetroviruses, and (-) strand RNA tospoviruses, have permitted to identify conserved sequence motifs involved in the virus movement process (Mushegian and Koonin 1993; Bouhida et al. 1993; Thomas and Moule 1995a, 1995b; Tzafrir et al. 1997; de Kochko et al. 1998). The figure below (pdb file 2ip5) shows a theoretical three-dimensional structure prediction of MOV, according to the work of Madhusudhan et al. (2006).
Related literature
Heinlein 2009; Taliansky and Torrance 2008; Taliansky et al. 2008; Waigmann and Heinlein 2007; Lucas 2006.
