Menu Tools: Functional analyses
[Menu Tools: Functional analyses
Functional Analyses overview
The horizontal main menu of GPRO implements a scroll down list to different GUIs allowing you to easily run from your PC comparative searches to the most common reference databases using a variety of software pipelines and free software tools installed in the GPRO computing server, or in your own server if you installed the GPRO protocol in your own cluster. As shown in Figure 8.1, GPRO gives you the option of performing automatic annotation using BLAST and HMM searches or INTERPROSCAN ((Quevillon et al. 2005). This section also allows you to make Ab initio gene predictions based on your query sequence using the software Augustus (Stanke et al. 2008).
|
Figure 8.1. Functional analysis options. |
BLAST and HMM searches
BLAST and HMM searches have been predominantly used to characterize the function of novel described sequences, either nucleotides or amino acids. In fact, the best hit using a BLAST search is sufficient to establish a relationship of homology between query and subject. Quality of annotation depends on the quality of the database used as the subject in the homology search. GPRO implements a pipeline installed for running the whole BLAST search process. You can manage the analysis and conditions from your PC but you launch the analysis at the server. This means that you must transfer the files from your PC to your user account at the computing cluster. In particular, this pipeline consists of the NCBI-BLAST package (Altschul et al.1997) and HMMER with a collection of scripts managed by a GUI presenting five tabs; “Format databases”, “BLAST analyses”; “HMM analyses”; “Process BLAST outputs” and “Process HMM outputs”.
Format databases
This tool is the first tab of the GUI for BLAST and HMM searches. It performs calls the “formatdb” script of the NCBI-BLAST package allowing you to easily give BLAST format to any fasta file created based on your own data or any RefSeq database downloaded from any internet source (Silva, Repbase, NCBI-NR, Uniprot, etc).
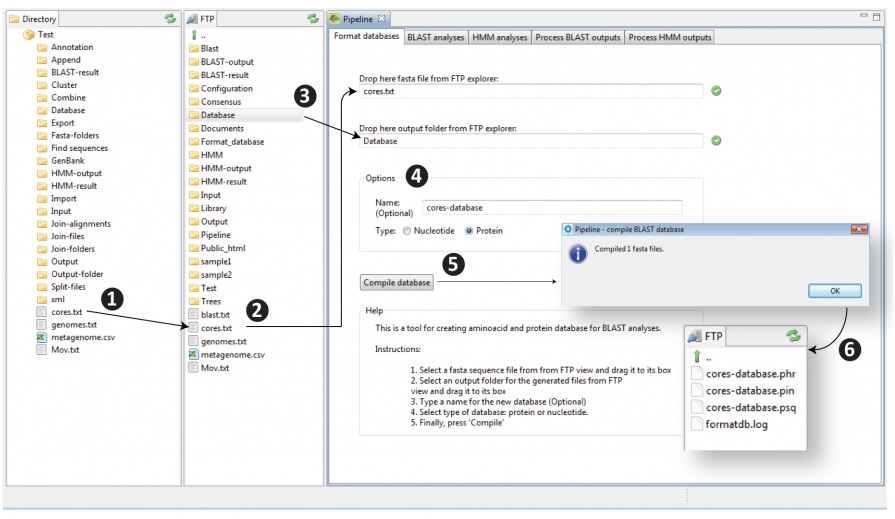
The procedure for formatting databases is shown in Figure 8.2 and detailed below according to the following steps:
- 1) Transfer the fasta file you want to format from your PC to your cluster account just dragging the file with the mouse from a place into the other (to the FTP explorer).
- 2) Then drag your input file with the mouse to the input box “Drop here fasta file from FTP explorer” displayed at the top of the GUI. If you did it successfully you will see a green icon at right to the box.
- 3) Drag the output folder (a new folder or an existing one) on which you want to store the BLAST database files (when your original file is BLAST formatted it will be in a set of three binary files) from the FTP to the output box (below to that for input). Again, if you did it successfully you will see a green icon at right to the box.
- 4) Give the database a name and select the type of database (nucleotide or amino acid)
- 5) Compile the database
- 6) The tool will automatically generate the three binary files (.phr; .pin; .psq) within the output folder. As previously noted, these three files constitute the formatted database recognized by the BLAST package compiled in GPRO.
|
Figure 8.2. Procedure for giving BLAST format to fasta files. In doing so you can create your own refseq databases in further BLAST comparisons using new sequences as queries to this refseq material |
BLAST analyses
“BLAST analyses” is the second tab of the GUI for BLAST and HMM searches. It calls the distinct tools (summarized in Table 1) provided by NCBI-BLAST package for similarity searches using both, nucleotide or protein fasta files as input queries to any (blast-formatted) refseq database. In other words, the tool allows you to manage in graphical mode the distinct BLAST programs and to characterize one or more fasta files with hundreds and thousands of sequences by simultaneous comparison with any a RefSeq database.
| BLASTP | Identifies protein queries or finds protein sequence homologs in a protein database |
| BLASTX | Finds similar proteins to translated DNA queries in a protein database |
| BLASTN | Identifies DNA queries or find DNA sequences similar to the queries |
| TBLASTN | Finds similar sequences to protein queries in a nucleotide database |
Figure 8.3 illustrates the typical steps for launching a BLAST analysis with GPRO. Following is a description for each step.
- 1) Upload the query file you want to analyze from your PC to the FTP explorer using the mouse as told in the previous section above.
- 2) Drag the query file from the FTP explorer to the input submission box.
- 3) Drag the folder containing your blast-formatted RefSeq databases to the database-input submission box. Then, the names of the distinct RefSeq databases available the submitted folder will be alphabetically listed in the box. The name highlighted in blue in the figure indicates the RefSeq database selected for being searched by the query. You can change the RefSeq database just scrolling the list and selecting that of your interest with the mouse. Note that the Figure emphasizes with a red circle, a link below the output folder box. This link gives you the option of using large RefSeq databases such as the NCBI NR, INTERPRO or others available for common use of all GPRO users that we pre-compile and update periodically because of their public nature and high size.
- 4) Select with the mouse an output folder for deposit your BLAST results and drag it from the FTP to the output submission box of the GUI.
- 5) Select the BLAST program (according Table 8.1 indications) and options (E-value cut-off value) to filter your search in box Option to the right and the top in the GUI.
- 6) Enter your e-mail address to receive notification when the job is complete. Bear in mind that searches performed with a certain number of query sequences, are usually computationally intense. GPRO performs the distinct search jobs in remote unattended mode. This means that you can launch the search and leave the tool. The analysis will keep running despite quitting the pipeline and closing GPRO.
- 7) Click run BLAST for running the analysis but wait until a message appears confirming that the analysis has been launched. Then you will see that info about your launched analysis appears in the summary at the bottom of the GUI. This information includes a column called “Actions” where you will see (indicated with a red arrow) a “red icon” accompanied by the word “Stop”. This button is for aborting the analysis at any moment if needed, so do not touch his button unless you want to do that.
|
Figure 8.3. Launching a BLAST search. Note that submission boxes and text areas include a check icon that can show two color states; green that means data successfully submitted/typed and red that will mean data not submitted or incorrectly typed. |
HMM analyses
“HMM analyses” corresponds with the third tab of the GUI for BLAST and HMM searches and provides an interface for performing HMM searches with HMMER based on two alternative options; HMMSCAN and HMMSEARCH. The first allows comparisons using a sequence fasta file as query to database of HMMs. The second performs comparisons using the HMM database as query to the sequence fasta file as subject. Figure 8.4 shows a screenshot of HMMER GUI although the procedure is, in overall terms, quite similar to that above described for BLAST searches. Just to note that you can create your own HMM databases using the HMMER server accessible via GUI in the Menu tab "Alignment analysis" or to download any RefSeq HMM databases from internet. It is also worth to remember that you can share pre-compiled databases in the repository system with other users if you are a user of the GPRO cluster. If you need to do so, just import the database of your interest by clicking on the link to the repository of pre-compiled databases highlighted in red within Figure 8.4. If not, let us to known and we will try to add it to the list of common resources.
|
Figure 8.4. HMM search screenshot |
Process BLAST outputs
Once a BLAST search is completed you will find an e-mail notification and the pipeline will report the result of the search in the folder selected as output folder. The BLAST result consists of a number of XML files, usually thousands because the search delivers one XML file for each sequence of the query file and all hits detected. XMLs must be thus processed in order to extract and export the obtained results in a single but interpretable annotation file. You can do this using the “Process BLAST outputs” script (Figure 8.5), which is the third tab of the GPRO GUI for BLAST and HMM searches.
Management of “Process BLAST outputs” is similar to that previously explained for other GUIs. Briefly, you must use the mouse to take the whole output folder into the input box labeled as “Drop here BLAST XML result folder” and then an additional empty folder into the box below “Drop here output folder. From that point in, the tools gives three options for processing the output XML files.
- The first option is a tool that retrieves and exports the BLAST results from all XML outputs to a single CSV file (the annotation file) consisting in as many rows as sequence queries and as many columns and annotation features. This CSV can be opened, visualized and managed via the annotation worksheet system of GPRO. You can also apply an e-value cutoff before running the script, take as many best hit per sequence query as desired, filter positional redundancies and append Gene Ontology (GO) annotation terms (Gene Ontology Consortium 2008). In doing so, the script will also include annotations based on the INTERPRO (Hunter et al. 2012) and KEGG (Nakaya 2013) systems.
- The second option generates the aforesaid CSV plus an additional FASTA database file with the FASTA header of the query sequences labeled with the BLAST results annotated according to the best BLAST hit information.
- The third option generates the CSV plus an additional FASTA database file with all the subject sequences detected by the queries but labeled with additional annotations according to the queries sequence fasta names.
Both the second and third options need additional information in order to append information to the fasta files to annotate in the analysis. When clicking on any of these options an additional form will appear to the left providing two additional utilities; “Fasta retrieval options” and “Additional retrieval option”.
“Fasta retrieval options” is needed in order to parse the files of which the information to append in the sequences to annotate in the fasta file generated in parallel to the CSV. Here, you can use a fasta file (usually the same query file) if you decide to create a database with subject sequences annotated on the basis of the queries, or alternatively use a BLAST compiled database if you choose to annotated the query sequences on the basis on the subject information. In the first case, just drag the query file to the box. In the second one, drag the folder you have for storing your RefSeq databases and then select from the list that will appear that used in the analysis.
“Additional retrieval functions” is a utility that allows you to decide if you want to export full sequences or just their alignment core of BLAST similarity. Furthermore, you can also ask the tool to retrieve the cores flanked by an additional number of nucleotides (in the case of DNA sequences) or residues (in the case of protein sequences) flanking them at both upstream and downstream.
|
Figure 8.5. Processing results from the XML outputs reported by the BLAST search. |
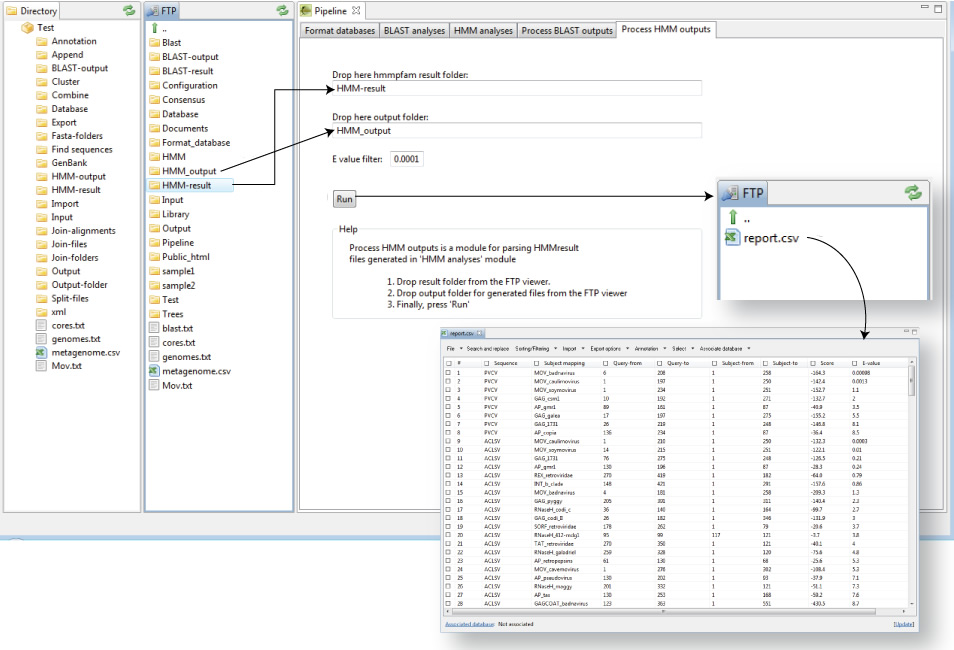
Process HMM outputs
“Process HMM outputs” is a script corresponding with the fifth tab of the GPRO pipeline GUI for BLAST and HMM searches. This script (Figure 8.6) allows you to export annotations and results from the output file generated by HMMER to a CSV file that can be opened, visualized and managed via the annotation worksheet system of GPRO. The procedure is very similar to that for processing BLAST outputs but with the difference of that HMMER does generates plain files as outputs instead of XMLs and also that “Process HMM” only permits annotation in a CSV (it does not generate additional fasta files).
|
Figure 8.6. Processing mapping results from HMM outputs |
InterproScan
INTERPROSCAN is a software package combining and different protein signature recognition methods native to the INTERPRO member databases (Hunter et al. 2012) into one resource with look up of the corresponding INTERPRO and GO annotation. For more details about INTERPROSCAN and INTERPRO databases, please refer to is web site and documentation at EMBL-EBI.
Running Interproscan via GPRO
By down scrolling the functional analysis tab of the main GPRO menu you can access a GUI for performing searches to any of all (or any) the INTERPRO database members using INTERPROSCAN. The GUI has two tabs, one for running INTEPROSCAN and the other for processing the XML output provided by this analysis. Figure 8.7 shows a screenshot of GUI section provided by GPRO when launching the search.
|
Figure 8.7. Running INTERPROSCAN. The procedure is quite similar to those previously explained in this section for functional analysis with the exception that you should to check the databases to which you want to perform your search. You can select all or any of them. |
Processing INTERPROSCAN outputs
Similarly to a BLAST search INTERPROSCAN generates distinct XML outputs as a result. By clicking on the second tab of the INTEPROSCAN GUI you will access to the interface of a script allowing you to obtain annotate your INTERPRO results and GO codes into a single CSV file similar to that provided the above mentioned script for processing BLAST outputs. This CSV consists of as many rows as sequence queries and as many columns and annotation features and can be opened, visualized and managed via the annotation worksheet system of GPRO. The procedure is similar to that when managing other GPRO GUIs.
|
Figure 8.8. Processing INTERPROSCAN XMLs into a single CSV. |
Augustus
AUGUSTUS is a program that makes ab initio prediction of genes in eukaryotic genomic sequences. AUGUSTUS can predict alternative splicing and alternative transcripts, as well as 5'UTR and 3'UTR including introns on species specific training sets. For more details about Augustus and the distinct species training sets supported by Augustus please refer to it web site
Running Augustus via GPRO
By down scrolling the functional analysis tab of the main GPRO horizontal menu you can access to a GUI to run Augustus. Figure 8.9 shows a screenshot of the GUI provided.
|
Figure 8.9. GUI provided by GPRO in order to run Augustus. The procedure is quite similar to all previously explained in this section for functional analysis with the exception of that you need to select the species training set on which you will base your prediction from the down scroll list available in the box species within the GUI (indicated with a red arrow) |