GIN/GINGER TRs and INTs
During the last years, the scientic literature has progressively accumulated sufficient evidence to convincingly argue a relationship to exist among the integrases (INTs) encoded by LTR retroelements, diverse host genes, and certain pools of transposase (TR) encoding transposons. To homogenize previous descriptions (Llorens & Marin 2001; Bao et al. 2010; Marin 2010; Marco & Marin 2009), we will collectivelly call these enzymes in this database as GIN/GINGER TRs and INTs. Included in the category of DDE TRS and INTs that comprises a variety of INT-TR-like enzymes, GIN/GINGER TRs ans INTs is here used to address those presenting significant similarity each to other (for more details about all DDE enzymes see the reviews of Nowotny 2009 and Hickman et al. 2010) or in other words an unequivocal and common evolutionary relationships. These pools are:
- The INTs coded by the LTR retroelements of eukaryotes
- The chromodomain-like TRs coded by the Maverick/Polinton giant transposons synthesizing transposons (Feschotte & Pritham 2005; Pritham et al. 2007; Kapitonov & Jurka 2006)
- The TR products encoded by the Tdd-like (Wells 1999; Glockner et al. 2001), the GINGER1 (Bao et al. 2010); the GINGER2 (Bao et al. 2010), the GINO (Marin 2010) and the GINA (Marin 2010) transposons of eukayrotes
- The IS3/IS481-like insertion sequences (ISs) described in prokaryotes and diverse protists (Fayet et al. 1990; Capy et al. 1997).
- The INT products encoded by the following host integrase genes of eukaryotes: FOB1 (Dlakic 2002); GIN1 (Llorens & Marin 2001); GIN2 (Bao et al. 2010; Marin 2010); CGIN1 (Marco & Marin 2009); the SCAN/KRAB carrying INT domain (Llorens et al. 2012, and references therein).
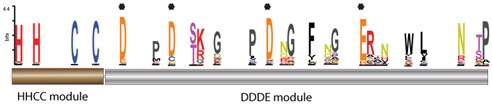
The aforesaid pools share an integrase core showing an N-terminal His-His-Cys-Cys (HHCC) module (not present in all pools) preceding a highly variable DDE module that exhibits, among other prominent amino acids, not three but four D, D, D, E residues (highlighted with asterisks in the figure below) that can vary depending on the pool.
The evolutionary relationships among the aforesaid pools challenge current concepts about selfish evolution as GIN/GINGER TRs and INTs probably constitute an ancient system of versatile and collaborative mobile genetic elements prone to recombine with other genetic elements to generate more complex selfish combinations, act as own transposons; and co-evolve with their genomic hosts to become conventional host genes. Within this system, LTR retroelement INTs and concretely those coded by Ty3/Gypsy LTR retroelements, constitute a recurrent source for the emergence of new transposons and host genes but also there is evidence of the domestication of other transposons such as GINGER1 and GINGER2.
Based on INT-like structural potential similarities, all GIN/GINGER INTs and TRs can be considered as representative members of the Retroviral Integrase Superfamily (Nowotny 2009) of nucleic acid-processing enzymes involved in; a) selfish evolution; b) replication and repair of DNA; c) recombination and gene fusion; d) RNA-mediated gene silencing; and e) oncogenesis.
