Element:ZFERV-2
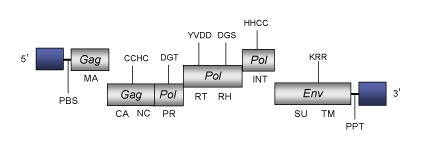
DescriptionZebrafish endogenous retrovirus from clone DKEY-26I24 (ZFERV-2) is counterpart of the ZERV element originally isolated from Zebrafish (Danio rerio) species (Shen et al. 2004). To date, ZFERVs have not been found in other teleost species, suggesting that this type of retrovirus may be limited to zebrafish (Shen et al. 2004). ZFERVs belong to genus Epsilonretrovirus within Retroviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 1 of the Retroviridae family. The genomic structure of ZFERV2 is 11.6 Kb in size including LTRs of 762 pb. The internal region of this retrovirus displays a Primer Binding Site (PBS), four Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Shen et al. 2004). The gag and pol ORFs are disrupted by a several stop codons and frameshiftings, but display the typical protein domains that are conserved in the Retroviridae family, including the gag, protease, reverse transcriptase, RNase H, and integrase domains (Shen et al. 2004). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||