Description
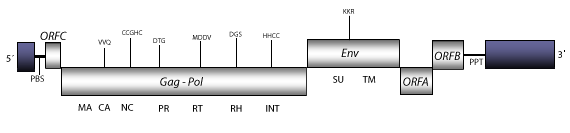
Walleye Dermal Sarcoma Virus (WDSV) is a complex fish retrovirus characterized in Stizostedion vitreum (walleye). WDSV is responsible for a skin tumour called walleye dermal sarcoma that produces seasonal lesions that appear in the winter and disappear in the summer (Bowser et al. 1988; Martineau et al. 1990; Martineau et al. 1992). Phylogenetically, WDSV is close to gammaretroviruses which usually are simple retroviruses. However, WDSV has several accessory genes and is therefore, a complex retrovirus constituting an own genus. The genomic structure of WDSV genome is 12.7 Kb size including pro-LTRs of 151 nt and 1.034 nt, respectively. The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNAHis, Open Reading Frames (ORFs) for gag-pol, and env genes characteristic of retroviruses, three accessory genes named OrfA (cicline-related), OrfB, and OrfC, this latter located between the U5 and gag gene, and also, a Polypurine Tract (PPT) adjacent to the 3´ LTR (Holzschu et al. 1995; Lapierre, Casey and Holzschu 1998 , Coffin, Hugges and Varmus, Map 13 1997).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|