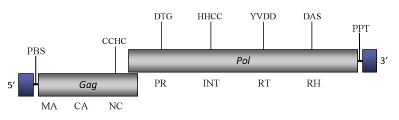
Element:Ty4
DescriptionTy4 is representative of one of the four LTR retrotransposon sets (Ty1, Ty2, Ty3 and Ty4) described in the genome of yeast Saccharomyces cerevisiae (Boeke and Sandmeyer 1991; Janetzky and Lehle 1992). It shows the greatest similarity with Ty2 and Ty1 elements and with them represents one of the members of Ty (Pseudovirus) clade, one of the three original genera on which the in-progrees Pseudoviridae classification is constituted (Boeke et al 2005). Ty4 genome is about of 6.2 Kb in size, including two 371-bp LTRs (Stucka et al. 1992; Janetzky and Lehle 1992). The LTRs flanked internal region contains two overlapping open reading frames (ORFs): the first ORF, TYA4, encodes a protein with the conserved motif (CCHC) found in the nucleic acid-binding gag-protein of retroviruses; the second ORF, TYB4, encodes a polyprotein showing domains with significant homology to the Ty1/Copia-like protease, integrase, reverse transcriptase, and RNase H (structurally arranged in this order) (Janetzky and Lehle 1992; Stucka et al. 1992). A potential primer binding site is located downstream to the 5’LTR, it differs from those of the other yeast Ty elements to be complementary to a tRNAAsn (Neuvéglise et al. 2002). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||