Description
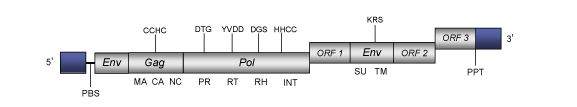
Snakehead fish retrovirus (SnRV) is a retrovirus originally isolated from a spontaneously productive infection of the SSN-1 cell line, derived from the Southeast Asian striped snakehead fish (Ophicephalus striatus) (Hart et al. 1996). SnRV within Retroviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 1 of the Retroviridae family. The genomic structure of SnRV is 10 Kb in length, including LTRs of 518 bp of sequence at each end of the proviral genome (Hart et al. 1996). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNAArg, four Open Reading Frames (ORFs) for gag, pol (PR, RT, RNase H, INT) and env (in two different ORFs located upstream of gag and downstream the ORF 1) genes characteristic of retroviruses, and with three different accessory genes of undescribed function (ORF 1, ORF 2 and ORF 3) as well as a Polypurine Tract (PPT) adjacent to the 3´LTR (Hart et al. 1996).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|