Element:RTBV
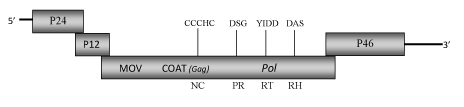
DescriptionThe plant pararetrovirus Rice tungro bacilliform virus (RTBV) is the only one reference species within the Tungrovirus genus (according to the International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). Diverse molecular studies performed with RTBV isolates from Philippine samples revealed that RTBV presents a bacilliform virion morphotype ranging 150-350 nm in lenght and 30-35 nm in width (Hibino et al. 1978). RTBVs usually encapsidate a circular dsDNA of about 8 Kb long showing two discontinuities in its two strands (Hay et al. 1991; Hull 1996 but can differ in size because of some nucleotides deletions/insertions (Hull 1996). For more details, see also ICTVdb). It is known that together with Rice tungro spherical virus (RTSV), RTBV constitutes a viral complex, which is responsible of "Rice tungro disease" the most important viral disease of rice (Oryza sativa) desctibed in South and Southeast Asia (Hibino et al. 1978; Hull 1996). RTBV is responsible for symptoms characterized by yellowing of leaves and stunting (Hibino et al. 1978) while RTSV is required for the transmission of both viruses by the leafhopper vector Nephotettix virescens (Hibino 1983a; 1983b). Endogenous RTBV-like (ERTBVs) sequences, rearranged and with no intact ORFs, have been found in the rice genome (Kunii et al. 2004). The RTBV genome contains four ORFs P24, P12, P194, P46 (Hay et al. 1991). The molecular function of P24 and P46 are yet under study (Hay et al. 1991; Hull 1996). Although the position of P46 is coincident with that of the typical translational activator (TAV) gene observed in other caulimoviruses, it is premature to postulate that the P46-product is involved in the control of RTBV expression (Hull 1996; Jacquot et al. 1997) as there is not apparent similarity between P46 and TAV products. P12 has no associated molecular function but it has been demonstrated that it displays the capacity to bind nucleic acids and that this property, already established for CSSV-ORF II protein (Jacquot et al. 1996), may be related with a sequence "PPKKGIKRKYPA" motif located at the C-terminus of p12. The largest ORF P194 encodes for a single large polyprotein containing the movement protein (MOV) as well as the coat protein, the protease, the reverse transcriptase and RNase H domains characteristic of retroelements and other caulimoviruses (Hay et al. 1991; Hull 1996). The COAT (gag-like) trait includes a large region (only observed in Badna- and Tungroviruses) that is rich in zinc finger (CCHC) arrays similar to those found in the LTR retroelement gag-nucleocapsids (Hull 1996; Bouhida et al. 1993; Llorens et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||