Description
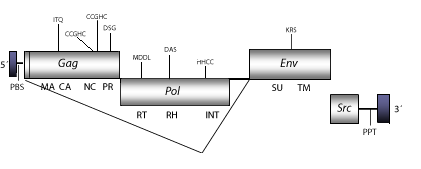
Rous Sarcoma Virus (RSV) is an avian oncogenic retrovirus, reported by Peyton Rous in 1911 as an infectious tumor agent inductor (Rous 1910). RSV has several variants, the Schmidt-Ruppin D strain has been taken as representative. The genomic structure of this clone is 9.3 Kb in size including pro-5´LTR and pro-3´LTR of 101 and 246 nt, respectively. The internal region displays a Primer Binding Site (PBS) complementary to a tRNATrp, Open Reading Frames (ORFs) for gag, pol, and env genes characteristic of retroviruses, an oncogen src, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Schwartz, Tizard and Gilbert 1983; Morris and Leis 1999). The src oncogen is evolutionarily related with a host-native proto-oncogene displayed in certain avian genomes, and codifies for a Tyrosine Kinase that binds phosphate groups to tyrosine residues in various host cell proteins (Steheling et al. 1976; Spector, Varmus and Bishop 1978; Bishop 1985).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|