Element:PyERV
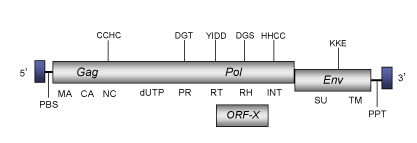
DescriptionPython endogenous retrovirus (PyERV) is a retrovirus originally isolated from Python species (Huder et al. 2002). PyERV belongs to genus Betaretrovirus within Retroviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 2 of the Retroviridae family. The genomic structure of PyERV is 7.0 Kb in size, including LTRs of 330 pb approximately (Huder et al. 2002). The internal region of this element displays a Primer Binding Site (PBS) complementary to a tRNATrp, two Open Reading Frames (ORFs) coding for two gag-pol and env polyproteins (gag-pol includes the dUTPase-protease characteristic of betaretroviruses) and a Polypurine Tract (PPT) adjacent to the 3´LTR (Huder et al. 2002). PyERV also shows an additional ORF (Huder et al. 2002) overlaping gag-pol, which was later identified as the typical ORFX of betaretroviruses (Llorens et al. 2008). For simplicity´s sake, the figure below shows an idealized single genomic organization) as the internal coding region of PyERV is disrupted by several stop codons and frameshiftings (see also, Huder et al. 2002). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||