Element:Opie-2
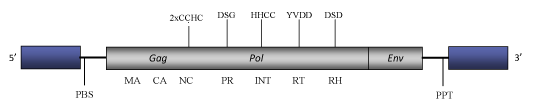
DescriptionOpie-2 is a Ty1/Copia LTR retrotransposon originally described in Zea mays (SanMiguel et al. 1996) and also found in rice (Oriza sativa, GenBank accession AC104473). Opie-2 belongs to SIRE a clade of Ty1/Copia plant LTR retrotransposons and potential retroviruses (Fauquet et al. 2005) evolutionarily related to another clade of plant Ty1/Copia LTR retrotransposons called Oryco (Llorens et al. 2009). SIRE clade gives the name to a genus within the current ICTV classification of the Ty1/Copia into three genera – Pseudovirus, Hemivirus and Sirevirus (Boeke et al. 2005). The Opie-2 full-length genome is 11.7 Kb in size, comprising both long LTRs of 1375 (5’) and 1348 nts (3’). The size difference between both LTRs is due to some nucleotides deletions within 3’LTR sequence. The internal region of this element contains a Primer Binding Site (PBS) complementary to a tRNAiMet, a coding region and a final Polypurine Tract (PPT) (SanMiguel et al. 1996). Although the internal coding region is disrupted by several stop codons and frameshiftings (for simplicity´s sake, the figure below shows an idealized single ORF organization), the distinct gag and pol domains typical of Ty1/Copia elements are well preserved in this element. Similarly to other Sire elements, Opie-2 contains a putative env-like gene at its C-terminal translated region (SanMiguel et al. 1996). In comparison to other Ty1/Copia elements, a particular feature of Sire-like elements meriting attention is their extremely large gag c-terminus consistent with the NC domain, which may display two or three CCHC arrays (Llorens et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||