Description
Murine Leukemia Virus (MuLV), occurring in mice is a C-type retrovirus that can be divided into four classes (Herr 1984; Stoye and Coffin 1987;O' Neill et al. 1985) according to the following host ranges:
- Ecotropic MLVs, which infect mouse cells and are horizontally and vertically transmitted in Mus musculus
- Polytropic MLVs, and modified polytropic MLVs, both usually isolated from leukemogenic tissues and capable of infecting mouse cells, and the cells of other organisms
- Xenotropic MLVs, unable to proliferate in mouse cells by infection but capable of infecting cells of other species
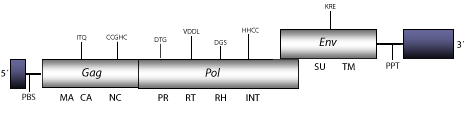
The xenotropic DG-75 MLV is taken as a representative, its genome is 8.2 Kb size including Pro-5´LTR and Pro-3´LTR of 143 and 471 nt, respectively. The internal region of this retrovirus displays two possible tRNA Primer Binding Sites (PBS) identified in genbank accession as Y=T and S=C for a tRNAThr; and Y=C and S=G for a tRNAGlu-2, a single Open Reading Frame (ORF) gag-pol, an ORF for env gene, and also a Polypurine Tract (PPT) adjacent to the 3´LTR (Raisch et al. 2003).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|