Element:Micropia
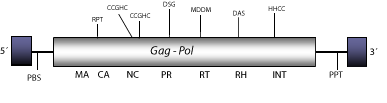
DescriptionMicropia is a LTR retrotransposon first described as a transcript expressed by fertility genes of chromosome Y of primary spermatocytes of Drosophila hydei (Henning et al. 1983; Huijser et al. 1988). Later, this element was also described in Drosophila melanogaster as a LTR retrotransposon combining structural features of DNA viruses, retroviruses and non-viral transposable elements (Lankenau et al. 1988). Micropia belongs to Micropia/mdg3 clade (Malik and Eickbush 1999). Micropia has several subtypes, the clone Dm11 of D. melanogaster is taken as a representative. The genomic structure of this clone is 5.5 Kb in size including LTRs of 480 nt. The internal region displays a Primer Binding Site (PBS) complementary to a tRNALeu, a single Open Reading Frame (ORF), which codifies for a gag-pol polyprotein, and a Polypurine Tract (PPT) adjacent to the 3´LTRs. Micropia also has a region of similarity to the class I major histocompatibility complex of mammals (Lankenau et al. 1988). This element also encodes an antisense RNA complementary to the Reverse Transcriptase_RNaseH region that seems to be involved in the control expression of transposon-encoded proteins in germ lines (Lankenau, Corces and Lankenau 1994). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||