Element:MMTV
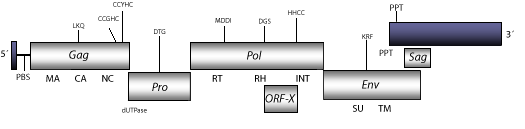
DescriptionMouse Mammary Tumor Viruses (MMTVs) are B-type retroviral inductors of mammary carcinogenesis and lymphoma in the T-cells of mice. This kind of retroviruses can be either exogenous (transmitted through milk) or endogenous (Bittner 1936; Callahan et al. 1982; Moore et al. 1987). MMTVs belong to the genus Betaretrovirus, which is comprised of both B- and D-type retroviruses. Interestingly, D-type betaretroviruses present a common surface receptor also used by the baboon and cat endogenous C-type gammaretroviruses (BaEVM, RD114), cellular infection by any of them can interfere with other D-type members, and also with BaEVM and RD114 (Chatterjee and Hunter 1980; Sommerfelt and Weiss 1990). This evidence seems to be related with the high similarity displayed between env polyproteins encoded by gammaretroviruses and D-type betaretroviruses, indicating that betaretroviruses and gammaretroviruses may have been involved in an ancient recombinatorial event (Sonigo et al. 1986; Fauquet et al. 2005) or that they share a common ancestor (see env tree in section "phylogenies"). The genomic structure of a typical MMTV is 8.8-9.9 Kb in size including pro-5´LTR of 135 nt and a long pro-3´LTR of 1.4 Kb. The internal region displays a Primer Binding Site (PBS) complementary to a tRNALys-3, Open Reading Frames (ORFs) for gag, dut/pro, pol, and env genes, an ORF for an accessory gene sag that encompasses the U3 3´LTR and env regions and codify for a 37-kDa type-II transmembrane glycoprotein known as superantigen (Sag), and a Polypurine Tract (PPT) adjacent to the 3´LTR (Moore et al. 1987; Acha-Orbea and Macdonald. 1995; Brandt-carlson and Butel 1991; Choi, Kappler and Marrack 1991; Choi, Marrack and Kappler 1992; Krummenacher and Diggelmann 1993; Yazdanbakhsh et al. 1993; Wrona et al. 1998; Elder et al. 1992; Payne and Elder 2001). We also have found that MMTV also presents a putative accessory gene Orf-x that is common among many other betaretroviruses, and that was originally described in JSRV betaretroviruses (Bai et al. 1999; Rosati et al. 2000). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||