Element:HIV-2
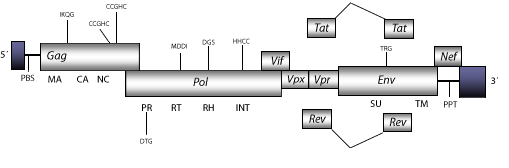
DescriptionHuman Immunodeficiency Viruses (HIVs) are complex retroviruses that induce Acquired Immunodeficiency syndrome (AIDS) in human beings. Two major types of HIV have been described; HIV-1, predominant world-wide and HIV-2, less virulent and prevalent in western Africa (Barre-Sinoussi et al. 1983; Gallo et al. 1984; Levy and Simabukuro 1985; Clavel et al. 1986). Both retroviruses are evolutionarily related to various Simian Immunodeficiency Viruses, nevertheless HIV-2 has not evolved directly from HIV-1, or vice versa (Hirsch et al. 1989; Guyader et al. 1986; Huet et al. 1990; Gao et al. 1999). There are 40 million people at present worldwide infected by Human Immunodeficiency Viruses -HIVs- (Prevalence map. 2006 Report on the global AIDS epidemic, UNAIDS, May 2006). At the genome structural level, HIV-2 is 9.6 Kb in size including LTRs of 1.029 nt (pro-5´LTR-R/U5 of 299 nt and pro-3´LTR-U3/R of 730 nt). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNALys3, Open Reading Frames (ORFs) for the gag, pol and env genes typical in retroviruses, tat and rev (both expanded in two exons) and vif, which are three accessory genes common in other lentiviruses, vpx, vpr and nef accessory genes specifical of primate lentiviruses, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Clavel et al. 1986; Guyader et al. 1987). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||