Element:HIV-1
DescriptionHuman Immunodeficiency Viruses (HIVs) are complex retroviruses responsible of inducing Acquired Immunodeficiency Syndrome (AIDS) in human beings. Two major types of HIV have been described; HIV-1, predominant world-wide, and HIV-2, less virulent and prevalent in western Africa (Barre-Sinoussi et al. 1983; Gallo et al. 1984; Levy and Simabukuro 1985; Clavel et al. 1986). Both retroviruses are evolutionary related to different simian immunodeficiency viruses, nevertheless, HIV-2 has not evolved directly from HIV-1, or vice versa (Hirsch et al. 1989; Guyader et al. 1986; Huet et al. 1990; Gao et al. 1999). HIV-1 comprises the following three groups: M group, O (outlier), and N (non-M, non-O). Group M is phylogenetically divided into 11 subtypes and responsible for the global HIV-1 pandemic. Recombination of M subtypes has resulted in the generation of multiple circulating recombinant forms (CRFs) consisting of mosaic lineages (Perrin, Kaiser, and Yerly 2003; Rambaut et al. 2004). Most subtypes and CRFs are represented in Africa reflecting the origin of the epidemy. There are 40 million people at present worldwide infected by Human Immunodeficiency Viruses (HIVs) (Prevalence map. 2006 Report on the global AIDS epidemic, UNAIDS, May 2006). Pathologically, an HIV infection presents the following three distinguishable clinical stages (Hazenberg et al. 2000):
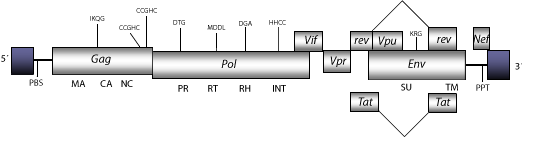
The HIV-1 particle displays a similar morphology to that of C-type retroviruses, but due to its conical form it has not yet received a morphological classification letter. The genomic structure of HIV-1 is 9.6 Kb in size including LTRs of 633-546 nt. Its internal region displays a Primer Binding Site (PBS) complementary to a tRNALys3; Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, tat and rev (both expanded in two exons) and vif, which are three accessory genes common in other lentiviruses, vpx, vpu and nef accessory genes specifical of primate lentiviruses, a Polypurine Tract (PPT) adjacent to the 3´LTR as well as another PPT found approximately in the central zone used as a secondary site in the beginning of the + DNA chain synthesis (Ratner et al. 1985). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||