Description
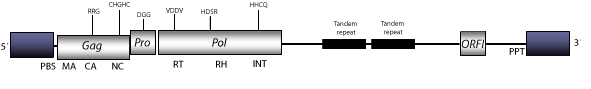
Grande is a LTR retroelement originally characterized in the genome of Zea diploperennis, and known to also be widely distributed in all Mais chromosomes (Aledo et al. 1995). This element belongs to a lineage of plant LTR retrotransposons (Tat), sibling of other lineage of putative plant endogenous retroviruses (Athila), providing evidence of a shared ancestral lineage originally described as class A by Wright and Voytas (1998). The genomic structure of Grande has several subtypes, subtype 1-4 is taken as representative. The genome structure of this element is 13.8 Kb in size including LTRs of 0.6 Kb. The internal region displays a Primer Binding Site (PBS) complementary to a tRNA Arg, three Open Reading Frames (ORFs) for gag, pro, and pol genes, and downstream to the integrase domain, a non-coding region of approximately 7 Kb in size, which contains two tandem repeats and an ORF that might be transcriptionally active in reverse frame, and also, a Polypurine Tract (PPT) adjacent to the 3´LTR (Martínez-Izquierdo, García-Martínez and Vicient, 1997; García-Martínez and Martínez-Izquierdo 2003).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|