Element:G-Rhodo
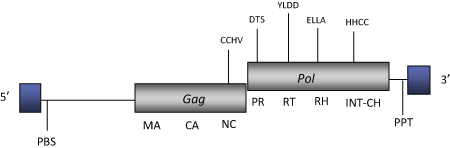
DescriptionG-Rhodo (Gypsy Rhodomonas) is a LTR retrotransposon characterized in the cryptomonad algae Rhodomonas salina (Khan et al. 2007). G-Rhodo belongs to the Chromoviridae branch, term suggested (Marín and Lloréns 2000) to describe the Ty3/Gypsy elements bearing chromodomain-integrases (Malik and Eickbush 1999). The chromovirus branch has been disclosed to be the most ancient phylogenetic pattern of Ty3/Gypsy retroelements (Gorinsek, Gubensek and Kordis 2004; 2005; Kordis 2005). The genomic structure of G-Rhodo is 8.7 Kb in size, including LTRs of 791-787 nt in size. The internal region displays a Primer Binding Site (PBS), two ORFs encoding for the typical gag and pol polyproteins of Ty3/Gypsy chromoviruses (including a chrodomomain at the C-terminus of the pol integrase domain) and a Polypurine Tract (PPT) adjacent to the 3´LTR (Khan et al. 2007). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||