Element:Fourf
Revision as of 10:02, 25 January 2010 by imported>Lcovelli
(diff) ← Older revision | Latest revision (diff) | Newer revision → (diff)
Description
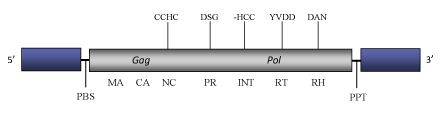
Fourf is a Ty1/Copia LTR retrotransposon described in Zea mays (SanMiguel et al. 1996). It is part of a complex nested structure involving at least nine other mobile genetic elements inserted in another Ty1/Copia LTR retrotransposon -RIRE1, which at the same time is inserted in another genetic element called Prem2-a (Fu et al. 2002). According to Llorens et al. 2009, Fourf belongs to Tork clade within Branch 2 of the Ty1/Copia LTR retrotransposon family. The genome of Fourf is about 7.0 Kb in size, including LTRs of 1.1-1.2 Kb (SanMiguel et al. 1996; Fu et al. 2002). The internal region of this element contains a Primer Binding Site (PBS) complementary to a tRNAiMet, a single long Open Reading Frame (ORF) of 1466 amino acids encoding for the gag and pol domains typical of Ty1/Copia LTR retrotransposons, and a Polypurine Tract (PPT) upstream to the 3´LTR (SanMiguel et al. 1996; Fu et al. 2002).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|