Element:DBV
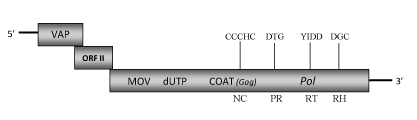
DescriptionYam (Dioscorea spp.),for their large underground tubers and aerial bulbils, is an important food in tropical and sub-tropical regions, particularly in West Africa and the South Pacific. A typical symptomatology, known as "internal brown spot" affecting tubers Yam, was found to be potentially associated to the presence of badnavirus-like particles (Harrison and Roberts 1973). Although several sequences have been identified in various Dioscorea species (Kenyon et al. 2008; Bousalem et al. 2009), to date only two -Dioscorea alata bacilliform virus (DaBV) and Dioscorea sansibarensis bacilliform virus (DsBV)- have been entirely characterized (Briddon et al. 1999; Seal and Muller 2007). The DBV sequence selected for our study is that isolated from yam Dioscorea sansibarensis species (DsBV). It is a member of the plant pararetrovirus genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). According to Llorens et al. 2009, DBV belongs to Badnavirus genus within Class 2 of the Caulimoviridae family. DsBV bacilliform particles of approximately 130- 30 nm in size, contain a circular double-stranded DNA genome 7261 bp long encoding for three Open reading frames (ORFs I, II, III) (Seal and Muller 2007). The putative ORF I protein is similar to those of the correspondent ORFs encoded by badnaviruses and showing sequence similarity to the Caulimoviridae virion associated proteins (VAPs). ORF II potentially encodes a protein of 14.1 kDa which function is still unknown. The amino acid sequence of ORF III contains the characteristic MOV, COAT, PR, RT and RH domains, including the large cysteine-rich region (only observed in Badna- and Tungroviruses) at the C-terminus of the COAT (gag) domain (Seal and Muller 2007; Llorens et al. 2009; Hull 1996; Bouhida et al. 1993). Interestingly, the DBV ORF III-polyprotein contains an additional dUTPase domain upstream to the COAT domain similar to that of several Retroviridae retroviruses (Elder et al. 1992) and Ty3/Gypsy LTR retrotransposons (Novikova and Blinov 2008). A similar dUTPase domain has also been found in the genome of other Badnavirus species, Taro bacilliform virus (TaBV). DBV is transmitted mechanically, as well as by the mealybug Planococcus citri from Dioscorea alata to some other Dioscorea species but, due to its low titer accumulation in yams, sometimes does not produce pronounced symptoms on leaves that only show mild deformation and veinal chlorosis (Phillips et al. 1999; Seal and Muller 2007). As flowering and seed-setting is rare, yam plants are usually vegetatively propagated by planting of small tubers or tuber pieces (Kenyon et al. 2008). The greatest risk of virus spread is that caused by vegetative propagation of infected material. Badnavirus-like particles, first reported in yams in the 1970s, were associated with a flexuous virus presents in D. alata and D. cayenensis species with "internal brown spot disease" (Harrison and Roberts 1973). Subsequently, serologically related bacilliform badnavirus particles have been detected in several other Dioscorea species of different geographical regions worldwide (Kenyon et al. 2008; Seal and Muller 2007; Phillips et al. 1999; Briddon et al. 1999). Kenyon et al. (2008) and Bousalem et al. (2009), show the existence of different Badnavirus-like species highly diverse and prevalent in yams of different geographical areas, some of which represent previously uncharacterised and unnamed species. In particular, three sequences (two groups) have been found highly divergent to the other new South Pacific yam Badnavirus groups, so has been supposed probably they represent either new Caulimoviridae genera or endogenous pararetrovirus sequences (Kenyon et al. 2008). Although the integration of badnavirus sequences into the genome of D. rotundata has been suggested, further analyses should be necessary to determine if these sequences are dead integrants or activatable sequences (Bousalem et al. 2009). Unfortunately to date, the lack of any sufficiently specific antisera to yam badnaviruses hinders to differentiate between viral episomal and integrated sequences (Bousalem et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||