Element:CaMV
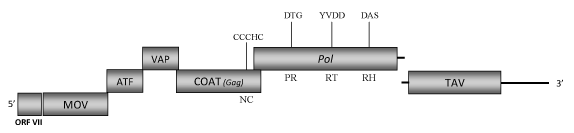
DescriptionCauliflower mosaic virus (CaMV) is the type species of the plant pararetroviral genus Caulimovirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). CaMV mainly infects a wide range of cruciferous plants, even if some strains are also known to infect Solanaceae species and both cruciferous and solanaceous plants (Piqué et al. 1995). The virus is transmitted mechanically as well as by aphid vectors. At least 27 aphid species are known as biological vectors of CaMV but the main ones in the field are Brevicoryne brassicae and Myzus persicae (Palacios et al. 2002). Several CaMV isolates are been described and the genomes of some have been sequenced completely (for more details see ICTVdb). The CaMV sequence selected for our study is that of the isolate "CaMV-Cabb-S" characterized in 1980s by Franck et al. (1980). Morphologically, CaMVs are spherical virions of approximately 50 nm in diameter containing a genomic circular double-stranded DNA (8024 bp long). The genome shows three single-stranded discontinuities, one in the (-) strand and two in the (+) strand as well as seven ORFs (I, II, III, IV, V, VI, VII) (Franck et al. 1980). ORF I encodes a protein of 327 amino acids (aa) associated to cell-to-cell movement (MOV) (Linstead et al. 1988). ORFs II and III encode for two proteins of 165 and 124 aa, which respectively correspond with the "aphid transmission factor" (ATF) and the "virion associated protein" (VAP) (Woolston et al. 1987; Hebrard et al. 2001; Leh et al. 1999). These two are both involved in the mechanisms of CaMV aphids transmission (Hoh et al. 2010; Palacios et al. 2002). ORF IV encodes the putative viral gag-like COAT protein and contains the typical caulimoviruses "RNA-binding"-associated motif, "C-X-C-X2-C-X4-H-X4-C" (Llorens et al. 2009; Hull 1996; Bouhida et al. 1993). ORF V is a pol polyprotein of 679 aa displaying the aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) typical domains. ORF VI encodes a protein of 520 aa long with a translational transactivator function (TAV) (Kobayashi and Hohn 2003). As other members of the genus Caulimovirus, CaMV encodes a small ORF VII upstream to the ORF I, whose translated protein has still an unknown function. The genome of CaMV also presents two intergenic regions, a large intergenic region (LIR) located between TAV and ORF VII, and a small intergenic region (SIR) between Pol and TAV. These regions contain two transcriptional promoter sequences that transcribe the whole genome of the virus (35S within the LIR) or only the ORF VI (19S within the SIR) (Hull 1984; Ow et al. 1987; Benfey and Chua 1990; Hohn and Fütterer 1992; Driesen et al. 1993). Their high expression activity make these promoters really useful tools for biotechnology applications and agricultural research (Fütterer et al. 1989; Hull et al. 2000; Ho and Cummins 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||