Element:CLNV
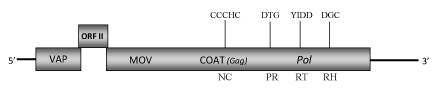
DescriptionAlthough cycad (Cycas revolute) is a plant species native to southern Japan, now is one of the most propagated and sold potted plant species in the world, growing outdoors in warm temperate and subtropical regions, or under glass in colder areas. Its common name "Sago Palm" has often originated a misunderstanding on its botanic taxonomy due to the fact that it is not a palm (Angiosperm, Monocotyledon) but it is a Gymnosperm. Recently has been identified the first plant pararetrovirus of Badnavirus genus in a Gymnosperm. It is infecting cycads from which derives its name Cycad leaf necrosis virus (CLNV) (Lockhart et al. 2008, unpublished). CLNV belongs to Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 2 of the Caulimoviridae family. Although CLNV is still an unclassificated badnavirus, it is probably the larger species sequenced among the genus because its bacilliform virions contain a circular dsDNA of about 9 Kb in size (9205 bp long) (Lockhart et al. 2008, unpublished). The genome of CLNV contains the three Open reading frames (ORFs I, II, III) characteristic of Badnavirus genus which potentially encoded proteins with molecular masses similar to those of the corresponding proteins of other badnavirus species. ORF I potentially encodes a 19.8 kDa protein showing sequence similarity to the Caulimoviridae virion associated proteins (VAPs) encoded by the corresponding ORFs I. The function of ORFII product (14.1 kDa) is still unknown even if a DNA-binding property has been hypothesized for the ORF II products of some badnaviruses (Borah et al. 2009; Jacquot et al. 1996). As reported for other badnaviruses, the ORF III product is a polyprotein with the typical cell-to-cell movement protein (MOV), viral coat protein (COAT), aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. It includes a "RNA-binding" motif sequence "C-X-C-X2-C-X4-H-X4-C" and a second large cysteine-rich region "C-X2-C-X11-C-X2-C-X4-C-X2-C" (only observed in Badna- and Tungroviruses) located at the C-terminus of the COAT (gag)-like region and similar to those identified in the nucleocapsid domains of LTR retroelements (Llorens et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||