Element:412
Revision as of 18:07, 10 November 2009 by imported>Gydbwiki
(diff) ← Older revision | Latest revision (diff) | Newer revision → (diff)
Description
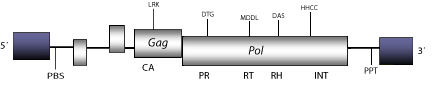
412 is a LTR retrotransposon described in Drosophilamelanogaster (Poter et al. 1979; Yuki et al. 1986a). In embryos of the Drosophila strain Df(3L)AC1 the expression of 412 is regulated by the homeotic genes abdominal-A and abdominal-B. This element provides thus a useful early marker for the development of the gonadal mesoderm in Drosophila embryos (Brookman et al. 1992). 412 is similar to mdg1 and belongs to 412/mdg1 clade (Malik and Eickbush1999). Phylogenetic analyses suggest that this clade is closely related to errantiviruses. The integrase product encoded by 412/mdg1 elements has been related to a single copy gene, which codifies for a putative integrase protein (GIN-1) in humans and other vertebrates (Lloréns and Marín 2001). The genomic structure of 412 is 6.9 Kb in size including LTRs of 310 nt. The internal region of this element displays a Primer Binding Site (PBS) complementary to a tRNA Lys, three Open Reading Frames (ORFs) coding for a gag precursor, another ORF for a pol precursor, and a Polypurine Tract (PPT) adjacent to the 3′LTR (Yuki et al. 1986b).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|