Caulimoviridae
Introduction
Caulimoviruses (Caulimoviridae) are DNA pararetroviruses that replicate in plants via a RNA intermediate evolved from LTR retroelements (for more details, see Bousalem et al. 2008). "Pararetrovirus" is the term introduced by Temin (1985) to define animal (Hepadnaviridae) and plant viruses (Caulimoviridae) that differ to retroviruses on the basis of their DNA genome and on their no regular integration into the host genome for replication. To date, it is known however that the genomic sequences of only a few plant pararetroviruses, thus as Petunia Vein Clearing Virus (PVCV) (Richert-Pöggeler and Shepherd 1997) and Banana Streak badnavirus (BSV) (Ndowora et al. 1999; Harper et al. 2005), are integrated into their host genomes, and that the integrated elements can give rise to episomal virosis (Ndowora et al. 1999). It is commonly assumed that these integrated sequences are relics of ancient infection events (Harper et al. 2005) or representative sequence intermediates between caulimoviruses and LTR retrotransposons (Bousalem et al. 2008; Llorens et al. 2009).
Virion morphology
Caulimoviruses usually form unenveloped virus particles which can be either bacilliform or isometric. As shown in the figure below, these are approximately 35-50 nm in diameter and 900nm in length (for the bacilliforms) or 45-50 nm in diameter and icosahedral symmetry (for the isometrics). The genome of caulimoviruses is a semi circular double-stranded DNA of about 6-8 kb in size, which is flanked by direct terminal repeats reiterated internally in an inverted form (for more details see Fauquet et al. 2005). The caulimoviral genome also has an intergenic poly (A) region (which may also be absent), and single-stranded discontinuities or gaps at specific sites of both strands (Harper et al. 2002).

Genomic structure
As noted in the section above, caulimoviruses have official taxonomical position within the viral nomenclature as pararetroviruses but although they are DNA viruses and do not have LTRs, they are evolutionarily related with eukaryotic LTR retroelements (particularly with those of the Ty3/Gypsy family, Llorens et al. 2009) on the basis of a gag-pol ancestor (on this topic, see also Koonin et al. 1991; Bousalem et al. 2008; Staginnus et al. 2009).
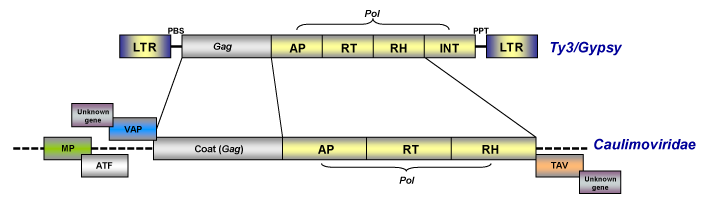
Caulimoviruses commonly show other genes such as those coding for the Movement (MP or MOV) protein, the Aphid Transmission Factor (ATF), the Virus Associated Protein (VAP) and the Transactivator/viroplasmin protein or Inclusion Body Matrix Protein (TAV or IBMp) as well as diverse additional genes (that vary depending on the lineage), which are necessary for their viral life cycle and transmission (for more details see Fauquet et al. 2005). Upon this, prior trends relate caulimoviruses to other virus systems based on the common share of the movement protein (Koonin et al. 1991), arguing that the most likely origin of caulimoviruses was chimeric (a hybrid between LTR retrotransposons and other RNA viruses).
Replication
Replication involves two phases: transcription of an RNA intermediate from the viral DNA in the nucleus and then reverse transcription of this RNA to rise dsDNA in the cytoplasm. The genome of pararetroviruses also contains a sequence complementary to a plant tRNAMet that corresponds to the initiation site of DNA replication. Usually this site is located inside or downstream of the large intergenic region (non-coding region) and is generally designated nucleotide 1 (de Kochko et al. 1998). In contrast to retroviruses, plant pararetroviruses do not require their integration into the host genome for their replication, therefore their genome does not encode the integrase protein. Although this is a characteristic of pararetroviruses, sequences from certain Caulimoviridae species, termed Endogenous Pararetroviruses (EPRVs, Staginnus et al. 2009) show a putative integrase domain similar to those found in retroviruses and retrotransposons of the Ty3/Gypsy family (Richert-Pöggeler and Shepherd 1997). In particular three viruses, BSV, PVCV and Tobacco vein clearing virus (TVCV) have been shown to have episomal infections associated with integrated sequences (Harper et al. 2002).
Biological distribution and host symptomatology
The natural hosts of Caulimoviridae species belong to the Kingdom Plantae (Angiosperms of Dicotyledonae and Monocotyledonae classes). Depending on the genera, the natural virus transmission could occurr via an insect vector (Hemiptera insects of the families Aleyrodidae Aphididae, Cicadellidae, Pseudococcidae) or by contact between plant hosts as well as by seeds or by pollen. Transmission can also be performed by techniques such as mechanical inoculation and grafting (for more details see Fauquet et al. 2005). The characteristic plant symptoms associated to caulimoviruses infection comprise: vein-clearing, banding mosaic, necrotic flecks, lines and chlorotic blotches, stunting, leaf curling, leaf malformation, twisting of leaflets, etc.
Evolutionary history and viral taxonomy
According to the International Committee on the Taxonomy of Viruses (ICTV, Fauquet et al. 2005), the Caulimoviridae can be taxonomically divided into six genera: Caulimovirus, Soymovirus, Cavemovirus, Tungrovirus, Badnavirus and Petuvirus. Phylogenetic reconstruction analyses based on pol collect these six genera into four classes: Class 1 includes the genera Caulimovirus and Soymovirus; Class 2 comprises Tungroviruses and Badnaviruses (the most abundant group of Caulimoviridae species); Class 3 represents the Cavemovirus genus; and finally Class 4 includes the Petunia Vein Clearing Virus (PVCV), which to date is the only known molecular species of Petuvirus genus (Llorens et al. 2009).
| Class | Host Phyla | Genus | Type species |
|---|---|---|---|
| Class 1 | Land plants (Viridiplantae) | Caulimovirus | Cauliflower mosaic virus (CaMV) |
| Class 1 | Land plants (Viridiplantae) | Soymovirus | Soybean chlorotic mottle virus (SbCMV) |
| Class 2 | Land plants (Viridiplantae) | Badnavirus | Commelina yellow mottle virus (CoYMV) or (ComYMV) |
| Class 2 | Land plants (Viridiplantae) | Tungrovirus | Rice tungro bacilliform virus (RTBV) |
| Class 3 | Land plants (Viridiplantae) | Cavemovirus | Cassava vein mosaic virus (CSVMV) |
| Class 4 | Land plants (Viridiplantae) | Petuvirus | Petunia vein clearing virus (PVCV) |
Class 1
Class 1 includes two genera – Caulimovirus and Soymovirus.
Caulimovirus
The species belonging to this genus form isometric particles with a diameter between 35 and 50 nm. The viral genome contains a single molecule of circular double-stranded DNA of about 8000 bp long that codes for 6 or 7 ORFs usually in the order addressed in the figure below, which is an idealized full-lenght consensus (for more details see Fauquet et al. 2005). Usually, the genomes of Caulimovirus species contain two major transcriptional promoter sequences: one located in the 3′-terminus of ORF VI (TAV) and extending into the large intergenic region that transcribes whole genome of the virus (a full-length transcript equivalent to CaMV 35S transcript); while the second one, situated at the 3′-terminus of ORF V (Pol) and extending into the small intergenic region between ORF V and VI, transcribes only the ORF VI, (a sub-genomic transcript equivalent to CaMV 19S transcript) (Bhattacharyya et al. 2002).
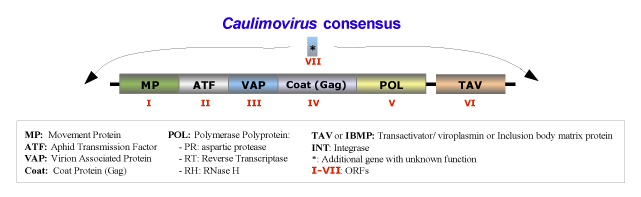
The natural plant hosts of the genus Caulimovirus are angiosperms of Dicotyledonae class. These viruses are usually transmitted in non- or semi-persistent manner by biological vectors, or via grafting, mechanical inoculation or by contact between hosts. The transmission vectors usually are insects of the order Hemiptera (Aphididae family), in which the virus does not replicate. Under experimental conditions, susceptible host species are found in the families Amaranthaceae, Caryophyllaceae, Chenopodiaceae, Compositae, Convolvulaceae, Cruciferae, Ericaceae, Euphorbiaceae, Leguminosae-Papilionoideae, Nyctaginaceae, Plantaginaceae, Ranunculaceae, Resedaceae, Rosaceae, Scrophulariaceae, Solanaceae. However some species of these families inoculated with virus do not show signs of susceptibility (i.e: Beta vulgaris, Brassica campestris Capsicum annuum, Chenopodium amaranticolor, Chenopodium quinoa, Cucumis sativus, Lycopersicon esculentum, Nicotiana benthamiana, Nicotiana clevelandii, Nicotiana tabacum, Petunia x hybrida, Phaseolus vulgaris, Pisum sativum, Spinacia oleracea, Vicia faba, etc).
According to the ICTV (Fauquet et al. 2005) the Caulimovirus genus contains viral species as diverse as the Cauliflower mosaic virus (CaMV); Carnation etched ring virus (CERV); Figwort mosaic virus (FMV); Dahlia mosaic virus(DMV); Strawberry vein banding virus (SVBV); Horseradish latent virus (HRLV); Mirabilis mosaic virus (MiMV); Thistle mottle virus (ThMoV); Aquilegia necrotic mosaic virus (ANMV, tentative species).
Soymovirus
Soymoviruses form round viral particles with icosahedral symmetry. The isometric capsid has a diameter of 42-50 nm and the viral genome contains a single molecule of double-stranded DNA of about 8200 bp long that forms an open circle (for more details see Fauquet et al. 2005). It contains 8 ORFs encoding for both structural and non-structural proteins (the figure below shows an idealized full-lenght consensus). Soymovirus genus differs from other caulimoviruses by the occurrence of three ORFs between ORF I and ORF IV instead of two (ORF II and III) that show little or no similarities to other caulimoviruses ORFs (Hasegawa et al. 1989; Mushegian et al. 1995). ORF VII reveals sequence similarity to the typical protease domain, it is not clear if this sequence is a functionally active protease additional to that found in pol but the feature merits further attention as it is taxonomically preserved in almost (but not all) soymoviruses. Moreover this genus differs in the location of the putative primer-binding site (PBS) that in the other caulimoviruses is usually located in the intergenic region, while in soymoviruses has been found within ORF A (PSCV) or ORF Ia (SbCMV) or between ORF A and B (BRRV) (Mushegian et al. 1995; Hasegawa et al. 1989; Glasheen et al. 2002).

The natural plant hosts of soymoviruses are angiosperms of Dicotyledonae class. Under experimental conditions susceptible host species are found in the family Leguminosae-Papilionoideae. Viruses are transmitted by mechanical inoculation, while they are not transmitted by seeds. According to the ICTV (Fauquet et al. 2005) the Soymovirus genus includes three species - Soybean chlorotic mottle virus (SbCMV), Peanut cholorotic streak virus (PCSV) and Blueberry red ringspot virus (BRRV) but although none tentative species has been yet reported, Stavolone et al.(2003) describe the new pararetrovirus Cestrum yellow leaf curling virus (CmYLCV) closely related to SbCMV.
Class 2
Class 2 consists of two other genera (Tungrovirus and Badnavirus) and it is the most abundant branch of Caulimoviridae elements.
This genus is characterized by species with bacilliform virions that have a length of 95-130 nm, or 60-900 nm as well as a width of 24-35 nm (for more details see Fauquet et al. 2005). The genome of badnaviruses usually contains a single molecule of dsDNA of about 7200-7600 bp long that forms an open circle interrupted by site-specific discontinuities and that could contain an intergenic poly (A) region (i.e CoYMV, Medberry et al. 1990). The virus genome usually codes for 3 ORFs, but in some cases can present one or four additional ORFs which functions are already under study. The largest ORF III polyprotein product contains the movement protein, the virus coat (gag) protein, the aspartic protease, the reverse transcriptase and the ribonuclease H domains (Medberry et al. 1990; Bouhida et al. 1993; Briddon et al. 1999). The COAT (gag) domain of badnaviral ORF III displays at the C-terminus a large region rich in zinc finger (CCHC) array duplications similarly to those of LTR retroelement nucleocapsids (Bouhida et al. 1993; Llorens et al. 2009). Additionally, some badnaviruses show an additional dUTPase domain upstream or downstream to the COAT domain within ORF III similar to that of several Retroviridae retroviruses (Elder et al. 1992) and several Ty3/Gypsy LTR retrotransposons (Novikova and Blinov 2008). The figure below shows an idealized Badnavirus full-lenght genome consensus.
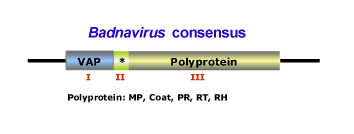
The plant hosts usually belong to the Dicotyledonae and Monocotyledonae classes. Depending on the Badnavirus species, the virus could be transmitted by mechanical inoculation, by grafting, by seeds or by pollen but not by contact between hosts. Badnaviruses can also be transmitted in a semi- or in a persistent manner via insect vectors of the order Hemiptera (Aleyrodidae, Aphididae, Cicadellidae and Pseudococcidae families). In addition, Banana streak virus (BSV) infections can arise in healthy plants from integrated sequences (as a result of the process of in vitro propagation) during tissue culture (Ndowora et al. 1999; Harper et al. 2002). According to the ICTV (Fauquet et al. 2005) the Badnavirus genus contains numerous species and tentative species, among which: Commelina yellow mottle virus (CoYMV or ComYMV) (type species in the genus), Aglaonema bacilliform virus (ABV), Banana streak virus (BSV), Cacao swollen shoot virus (CSSV), Citrus yellow mosaic virus (CMBV), Dioscorea alata bacilliform virus (DaBV), Kalanchoe top-spotting virus (KTSV), Sugarcane bacilliform virus (ScBV), have been described together with tentative species such as Pineapple bacilliform virus (PBV), Yucca bacilliform virus (YBV), Stilbocarpa mosaic bacilliform virus (SMBV).
Tungrovirus
Only one species has been referred to Tungrovirus genus: Rice tungro bacilliform virus (RTBV). The species consists of bacilliform particles long 110-400 nm and with a width of 30-35 nm that encapsidate a circular dsDNA of about 8000 bp long (for more details see Fauquet et al. 2005). The genome contains four ORFs that potentially encode for four proteins (P24, P12, P194, P46 respectively, Hay et al. 1991). The largest ORF (III) encodes for a polyprotein containing the movement protein as well as the coat protein, the protease, the reverse transcriptase and RNase H domains characteristic of retroelements. The functions of the three other tungrovirus ORFs are unknown or not well yet demonstrated (Hay et al. 1991; Hull 1996), (figure show the genome organization of the type species Rice tungro bacilliform virus, RTBV).

The plant hosts of RTBV belong to the Angiosperms of Monocotyledonae class. The virus is transmitted by insect vectors of the order Hemiptera, family Cicadellidae in a semi-persistent manner and requiring the presence of the Rice tungro spherical virus (RTSV) helper virus for transmission. RTBV and RTSV are both responsible for the "Rice tungro virus disease", one of the major causes to rice loss production in South and Southeast Asia (Hull 1996). Endogenous RTBV-like sequences (ERTBVs) have been found in the rice genome (Kunii et al. 2004). Although they contain rearranged structures and no intact ORFs, the comparisons of their DNA and amino acid sequences suggested their closely relationship to RTBV and the possible role of these integrated sequences against the related viral disease (Kunii et al. 2004).
Class 3
Class 3 represents the genus Cavemovirus.
Cavemovirus
This genus consists of two viral species - Cassava vein mosaic virus (CsVMV) and Tobacco vein clearing virus (TVCV) - according to the ICTV (Fauquet et al. 2005) and supported by phylogenetic analyses (Llorens et al. 2009). These form round-to-elongated capsids with icosahedral symmetry and length and width of 95-130 nm (or 60-900 nm) and 24-35 nm, respectively (for more details see Fauquet et al. 2005). The viral genome usually consists of a single molecule of circular dsDNA of about 7.7-8.1 Kb long that encodes for 4 or 5 ORFs (the figure below shows an idealized consensus of the cavemoviruses genome organization).

Cavemoviruses differ from other Caulimoviridae genera, in the order of their coat and movement domains (de Kochko et al. 1998; Calvert et al. 1995; Lockhart et al. 2000). In cavemoviruses, mov is downstream to coat. Additionally, CSVMV apparently codes for an extra protease domain, which is found within ORF I and upstream to the conventional pol protease domain. This feature is found in other Caulimoviridae species but not in TVCV. On the other hand, an interesting observation has been proposed by Lockhart et al. (2000) who suggest that the episomal form of TVCV found into the infected hybrid tobacco species N. edwardsonii (N. clevelandii x N. glutinosa) probably arises from integrated pararetroviral sequences present in the host plant genome and that they are inherited from the male parent (N. glutinosa).
Class 4
This class describes the genus Petuvirus, which falls in the deepest position of the Caulimoviridae phylogeny .
Petuvirus
This is a genus to date composed only of a single sequence representative, the Petunia vein clearing virus (PVCV), which might constitute the most basal position in the Caulimoviridae phylogeny (Bousalem et al. 2008; Llorens et al. 2009). This viral species consists of unenveloped isometric particles of 43-46 nm in diameter (for more details see Fauquet et al. 2005). The genome is not segmented and consists of a molecule of circular dsDNA 7206 bp long, organized into a large ORF containing the movement protein typical of caulimoviruses, a putative [HHCC and DD(35)E] integrase similar to those coded by LTR retrotransposons, and the typical coat (gag) and pol (protease-reverse transcriptase-RNaseH) domains (Richert-Pöggeler and Shepherd 1997; Harper et al. 2002). The presence of a probable integrase function in the genome of this virus suggests that PVCV exists as a viral retroelement that may also has the potential to transpose in Petunia (Richert-Pöggeler and Shepherd 1997).

The plant hosts of PVCV belong to Solanaceae family (Angiosperms of Dicotyledonae class). The virus has been found in Petunia x hybrida (garden petunia) in which the characteristic symptoms are vein clearing and leaf malformation, and it is highly transmitted by seed and grafting but not by mechanical inoculation or by insect (aphid) vectors (Harper et al. 2002).
