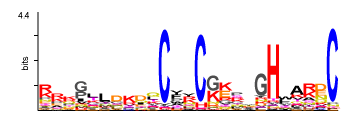Gag
Gag-like genes harbored by eukaryotic LTR retroelements encode for a gag precursor, which is usually processed by protease into matrix (MA), capsid (CA), and nucleocapsid (NC) proteins (Wills and Craven 1991).
- MA is necessary for membrane targeting of gag polyprotein and for capsid assembly.
- CA forms the prominent hydrophobic core of the virion (Stromberg et al. 1974).
- NC is involved in RNA packaging through recognition of a specific region of the viral genome called Ψ (Gorelick et al. 1988; Katz and Jentoft 1989; Khan and Giedroc 1992; Berkowitz, Fisher and Goff 1996).
The best-conserved part of the gag polyprotein is the CA-like major homology region (MHR), which usually displays a central QG-X2-E-X5-F-X2-L-X2-H motif implicated in the transposition (Patarca and Haseltine 1985; Wills and Craven 1991;Orlinsky et al. 1996; Nakayashiki et al. 2001). A second similarity within gag polyproteins is found in the C-terminus of the NC as a Cys-X2-Cys-X4-His-X4-Cys (CCHC) motif (as shown in the figure below), which may be absent or found one, two, or three times duplicated depending on the viral species (Berg 1986; Covey 1986; Green and Berg 1989; Llorens et al. 2009). CCHC arrays have been found to be critical for many steps in the viral life cycle, and several studies have shown they are involved in virion assembly, RNA packaging, reverse transcription, and integration processes (Buckman, Bosche, and Gorelick 2003 and therein references ). Each CCHC motif coordinates a zinc atom (Bess et al. 1992; Summers et al. 1992).
It is not yet clear which sequence functionally substitutes the CCHC array in those LTR retroelements in which it is absent. However, a glycine-arginine-rich zone near the C-terminal end of NCs encoded by spumaretroviruses, has been shown to have nucleic-acid-binding properties (Yu et al. 1996).