Retroelements
Introduction
Retroelements are mobile genetic elements (MGEs) that retrotranspose via a RNA intermediate that is reverse-transcribed to DNA by the encoded reverse transcriptase and integrated into a new location within the host genome by an integrase enzyme. They have been found among different organisms from bacteria to humans and often constitute a significant part of genomes, particularly in higher plants and fungi. Various retroelements with different gene organizations and replicative mechanisms have evolved in the course of evolution. Although in most cases they have no effect on the host organism, there are many examples of mutations caused by retroelements resulting in various diseases. However co-adaptation has led in some cases to the use of retroelements for essential and beneficial host functions.
Retroelements (retrotransposons and retroviruses) can currently be divided into four systems or groups commonly known as the long terminal repeat (LTR) retroelements, the non-LTR retroelements, the tyrosine recombinase (YR) retroelements and the Penelope retrotransposons (Eickbush and Jamburuthugoda 2008).
LTR retroelements
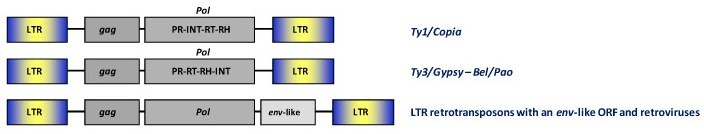
These include the broad range of LTR retrotransposons and retroviruses circulating in plants, fungi and animals. A full-lenght consensus LTR retroelement genome is characterized by an internal translating region (gag and pol genes, and env gene when is present) flanked by long terminal repeats (LTRs). They can be classified into four major groups or families based on sequence similarity and other features known as the Ty1/Copia, the Ty3/Gypsy, the Bel/Pao, and the Retroviridae families (for more details, see LTR retroelements).
Non-LTR retroelements
These constitute a system of retrotransposons widely distributed in eukaryotes, which do not present LTRs or terminal repeats; non-LTR retrotransposons end most frequently with a poly(A) tail at their 3′ end, while their 5′ end often contains variable deletions (5′ truncations). Depending on their capability to transpose or not autonomously these elements are classified as autonomous and non autonomous retroelements, respectively.
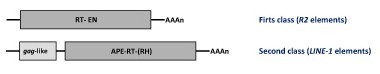
Autonomous non-LTR retroelements. On the basis of their molecular structures the autonomous non-LTR retroelements have been grouped in two major classes:
R2 elements : these constitute one of the most studied families of non-LTR retroelements (Eickbush 2002). They encode for a single ORF with a central RT domain and an endonuclease (EN) conserved domain at the C-terminus.
Long INterspersed repetitive Elements (LINEs) : these are a family of 6-8 kb long elements encoding for two ORFs. The first ORF shows similarity to retroviral gags. The second ORF encodes for a pol polyprotein displaying RT and apurinic-apyrimidinic endonuclease (APE) domains. However, some lineages additionally include a protein domain of unknown function at the C-terminus and in other cases an RNase H (RH) downstream of the RT domain as usual in LTR retrotransposons (Eickbush and Jamburuthugoda 2008). LINEs are widespread in mammals (Moran and Gilbert 2002).
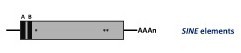
Non autonomous non-LTR retroelements. These are DNA sequences of 80 to 630 bp known as Short INterspersed repetitive Elements (SINEs). They represent reverse-transcribed RNA molecules originally transcribed by RNA polymerase III into tRNA, rRNA, and other small nuclear RNAs (Malik and Eickbush 1998). Like the LINEs, the SINEs end by a poly(A) tail or by A- or T-rich sequences, their 5’ and 3’ ends reveal similarities to tRNA genes (or, as shown for some animal SINEs, to 7SL RNA gene) and to the 3’ end of LINEs, respectively (Oshima et al. 1996). Two conserved sequence motifs found in the SINE tRNA-like part, called box A and box B, show homology to RNA polymerase III promoters. SINEs do not encode their own reverse transcriptase and are therefore unable to transpose autonomously. For this reason it has been proposed that SINEs use the enzymatic machinery of LINEs for their retrotransposition (Luan et al. 1993; Wallace et al. 2008; Kroutter et al. 2009). LINEs and SINEs elements have been not only described in the genomes of animals but also in many species across the Plantae Kingdom (Schmidt 1999).
Tyrosine recombinase (YR) retroelements
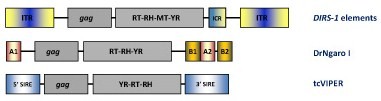
YR retroelements have been found in plants, protists, fungi, as well as a variety of animals including vertebrates, echinoderms and nematodes. The "gag-RT-RNaseH" genome organization of YR retroelements is similar to that of LTR retroelements but differ in the fact that YR retroelements lack the PR and usually show a tyrosine recombinase (instead of INT), which is typically involved in site-specific recombinations between similar or identical DNA sequences. YR retroelements can be divided into three families DIRS, Ngaro and VIPER (Goodwin et al. 2004; Goodwin and Poulter 2004; Vazquez et al. 2000; Lorenzi et al. 2006). The typical structure of DIRS elements contains inverted terminal repeats (ITRs), internal ORFs and an internal complementary region (ICR) derived from the duplication of flanking ITR sequences. DIRS elements differ from the two other YR-like families in the presence of a conserved methyltransferase (MT) domain at C-terminal to RT/RH that is similar to those MTs encoded by various bacteriophages (Goodwin and Poulter 2004). Ngaro elements show an ORF organization similar to that of DIRS elements but differ in the orientation of flanking repeats. While DIRS elements are delimited by inverted repeats, Ngaro elements are flanked by direct repeats (referred as A1, A2, B1 and B2). Ngaro elements have been found in different organisms as the zebrafish Danio rerio (DrNgaro1), as well as in fungi and in echinoderms (Goodwin and Poulter 2004). In turn, VIPER (Vestigial interposed retroelement) elements have been described in the genomes of trypanosome protozoan parasites. As an example, the figure below also shows the genomic organization of tcVIPER, an element described in the genome of T. cruzi. This element contains a coding internal region flanked by a lineage of SINEs (called SIRE) also found in T. cruzi genome (Lorenzi et al. 2006; Vazquez et al. 2000).
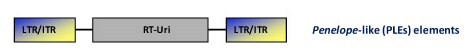
Penelope retrotransposons (PLEs)
This is a family of retrotransposons described in many animal genomes (more than 80 species belonging to at least 10 animal phyla), protists, fungi, and plants (Arkhipova 2006). Their genome structure contains apparent LTRs that may be in either direct or inverted orientations flanking a coding region with RT and EN domains (i.e. a pol polyprotein domain). Phylogenetic reconstruction analysis indicate that PLEs-like RTs are closer to telomerase RTs (TERTs) than to any other characterized RTs (Arkhipova et al. 2003). In turn, PLE-like ENs are related with the intron-encoded endonucleases and the bacterial repair endonuclease UvrC, both belonging to the Uri family of ENs (Pyatkov et al. 2004). Studies realized with the fly Drosophila virilis and bdelloid rotifer organisms revealed that the majority of PLEs in these species contain spliceosomal introns (Arkhipova et al. 2003). The peculiar structural organization of PLE elements, their ability to retain introns during transposition, and the distinct placement in the phylogeny of retroelements, suggest that PLEs constitute an ancient class of retroelements (Arkhipova 2006; Schostak et al. 2008).