Description
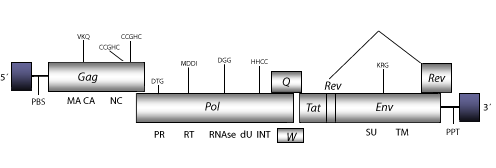
Maedi Visna Virus (VMV) is a lentivirus with antigenic variation and described in sheep and goat genomes (Sonigo et al. 1985; Braun, Clements and Gonda 1987; Clements et al. 1988). The genomic structure of a typical VMV retrovirus is 9.2 Kb including LTRs of 510 nt (pro 5´LTR-R/U5 and pro 3´LTR-U3/R of 160 and 350 nt, respectively). The internal region displays a Primer Binding Site (PBS) complementary to a tRNALys, Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, tat and rev and orfq (vif or sor), which are three accessory genes common in other lentiviruses, an orfw also displayed but corrupted in other sheep lentiviruses, and adjacent to the 3´LTR, a Polypurine Tract (PPT) common to other LTR retroelements (Sonigo et al. 1985). As other non-primate lentiviruses, VMV displays between the RNase H and INT domains, an ORF coding for a dUTPase (dU) similar to that observed in betaretroviruses, N-terminal to the PR domain (Elder et al. 1992; Payne and Elder 2001 and references therein).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|