Mouse functions and tricks
Mouse functions and tricks
How to deal with GPRO depending on the GUI
GPRO is very easy and simple to use. Almost all file and folder actions to perform on the GUI can be executed by selecting with the mouse the file / folder to use and then moving it with the mouse to the corresponding dialog box of the GUI being used. There are also functions accessible by right-clicking the mouse that can vary depending on the GUI being used. As all these utilities might not immediate for a user when using GPRO for the first times, this section is oriented to show you a perspective about the availability and utility of these tricks depending on the used GUI.
At directory space
The directory space of GPRO is organized following the typical tree-file dynamic hierarchy. You can thus navigate the folders containing folders and files etc. Figure 4.1 presents an idealized perspective of all right click utilities of the mouse. In doing so on the directory space, a dialog will appear offering you diverse actions. You can create a new file/folder, open a worksheet, and use the editors. You can also cut, copy, paste, delete and rename files etc via this dialog. Finally, at the bottom of this dialog, you will also access to a parser for extracting data from files formatted as GenBank accessions (indicated with a left-oriented arrow). This parser (indicated by a right-oriented arrow) let you to extract and create a sub-database with sequences and symbols and other coding or not coding features if they are annotated in the accession file. You can also select and extract sequence frames delimited by particular starting and ending positions within the accession. The tool is really useful in order to manage large files such as for instance, a full-length human chromosome accession, etc.
| Figure 4.1. Right-click utilities when positioning the mouse on the directory space |
About the database editor and fasta explorer
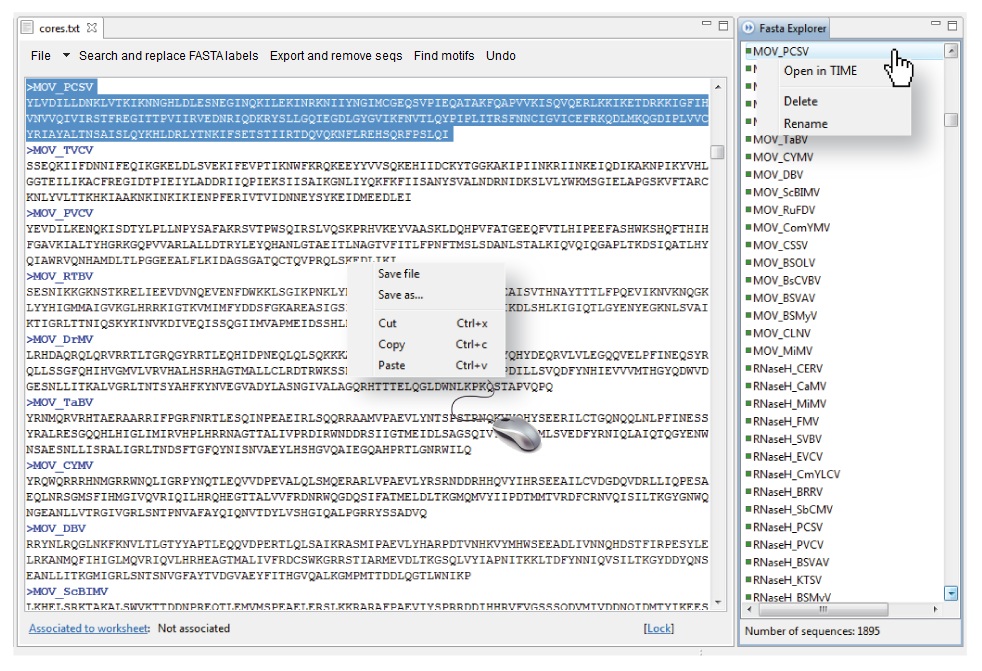
By positioning and right-clicking the mouse on the Database editor, this contextual menu provides distinct functions for editing (cut, copy and paste) contents. In addition, by clicking on any sequence name summarized in the FASTA Explorer, the database editor browses the edited file to reach particular sequences, which appear highlighted in blue when selected in the Fasta Explorer (Figure 4.2). The latter also allows opening of selected sequences in the TIME sequence editor, and also allows you to delete and rename selected sequences.
| Figure 4.2. Database editor contextual menu and fasta explorer. |
Dealing with the worksheet
The worksheet system is an excel-like template of rows and columns with which manage a variety of annotation, analyses and data mining actions. Rows represent sequences and columns the distant annotation features (we will see this more in deep in subsequent sections of this manual). The worksheet can be handled via the worksheet-menu or via mouse-dependent actions, which are additional to those available in the worksheet menu. Figure 4.3 shows an idealization of all the available mouse actions. For instance, to export a sub-annotation of your project into a new database you can select rows (sequences) and columns (some of their attributes) by manually clicking the checkboxes of each row and column of interest. You can also positionally switch columns by just selecting and dragging one of them from one position into another. More, all worksheet cells (including column headers) can be edited. By right clicking the mouse a little dialog will appear with two options “Columns” and “Rows”. The first one offers you a sub-dialog with additional actions for adding, selecting, removing renaming or joining and splitting columns. The second one drives to another sub-dialog permitting the same actions in regards of rows but letting you additional options. You can place the mouse in one row and them make right click > Rows > Show BLAST multihit. This action allows you to visualize the set of alternative best BLAST hits (if you are dealing with a multihit CSV file) for you sequence in the selected sequence and switch it (if appropriate) by another with an alternative description. Do not worry if you are unclear about the details and functionalities of this option, we will explain it more in depth in subsequent sections of this manual. Similarly, you can select the best isoforms among a selection of sequences annotated according to the same Genbank accession or any other feature. Again, do not worry about this, as we will explain it later with much more details. Finally, by double clicking on any row, such a row will appear below to the worksheet as a notebook for detailed editing (as shown in the figure).
| Figure 4.3. Worksheet functions and utilities dependent on mouse actions |
FTP explorer and pipeline GUIs
If you launch, via the main menu, any pipeline or server a FTP protocol called as FTP explorer, which is linked with your user account at the remote computing cluster will automatically appear right to the directory together with a GUI that varies depending on the pipeline. As shown in Figure 4.4, if you want to run a pipeline and/or server analysis your data files must be transferred from your PC to your user server account. You can do that by just simply dragging files with the mouse from your GPRO directory to your user account. Meanwhile the file transfer is under progression, a parallel pop-up will appear in order to inform you about the status of the process. Once your files are in your user server account you can launch the tool by selecting with the mouse the file (and other material if requested) to analyze and dragging it to the dialog box to which it corresponds to.
| Figure 4.4. Transferring files from your PC to the remote computing cluster via FTP and actions to launch the pipeline and server tools via GUI |
Working with management and data mining utilities
GPRO provides you a variety of tools for data mining and database management which are run in your PC via the main menu. As shown in Figure 4.5 these tools also have particular GUIs, which can be launched using the mouse with selecting and dragging actions in identical way to those examples summarized above in the previous examples.
| Figure 4.5. Management and data mining utilities dependent on mouse select and drag actions. |