Mobilomics-acyrthosiphon pisum-Comments and Discussion
Commments and Discussion
In the screening performed for the first release of the genome of Acyrthosiphon pisum (IAGC 2010), 549 LTR retroelement-like sequences including full-length and fragmented elements were identified and annotated. A summary of the features is presented in Table 1.
| Classification and Taxonomy | Total LTR retroelement-like features | Full-length genomes | Potentially active | Retroviruses |
|---|---|---|---|---|
| Ty3/Gypsy | 282 | 121 | 69 | 8 |
| Bel/Pao | 178 | 86 | 48 | 2 |
| Ty1/Copia | 12 | 6 | 3 | 0 |
| Related transposases | 35 | 0 | 0 | 0 |
| Unclassified sequences | 42 | 9 | 0 | 0 |
| Total | 549 | 222 | 120 | 10 |
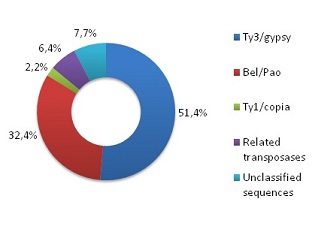
Almost all the annotated sequences correspond to LTR retroelement-like sequences belonging to the Ty3/Gypsy, Bel/Pao and Ty1/Copia families. In addition, the aphid genome contained many ORFs encoding for two pools of transposases (TRs) belonging to GINGER1 and GINGER2 DNA transposons (Bao et al. 2010) and other TRs related to those encoded by the Maverick/Polinton transposons (Pritham et al. 2007; Kapitonov & Jurka 2006). These were annotated as CIN1 (chromodomain-INTs type 1) because of their chromodomain (CHR), following the INT core at the C-terminal. These features were similar to the LTR retroelement used in queries during the processes of curation (see methods) as they share evolutionary history with the INTs coded by LTR retroelements. In addition, a number of sequences (unclassified) that were similar to our queries, but could not be assigned taxonomy levels on the basis of LTR retroelements, were annotated. As demonstrated in Figure 1, Ty3/Gypsy is the most representative family (51.4%) in the aphid genome, followed by the Bel/Pao family (32.4%). Unclassified features represent 7.7% of annotated sequences, the GINGER-like and the Maverick/Polinton TRs represent 6.4%, and the Ty1/Copia ORFs 2.2% of annotated sequences.
Of the 549 annotated sequences, 182 correspond to distinct LTR retroelement-like features scattered throughout the host genome as “Solo-genes”. Almost all of these are remnants of ancient full-length LTR retroelements fragmented because of distinct recombination processes and accumulation of indels during evolution (Petrov 2002). Table 2 summarizes the most relevant Solo-genes identified in the pea aphid genome, classified according to their gag, pol or env identity. The majority of these features are probably active genes, as they present with a high degree of sequence preservation and no stop codons. Among these, Solo-INTs were the most abundant features.
| Classification and Taxonomy | gag | pol | env | |||
|---|---|---|---|---|---|---|
| solo-gag | solo-PR | solo-RT | solo-RNAse H | solo-INT | solo-env | |
| Ty3/Gypsy | 9 | 11 | 10 | 1 | 25 | 1 |
| Bel/Pao | 14 | 4 | 8 | 1 | 18 | 1 |
| Ty1/Copia | 3 | 0 | 1 | 2 | 1 | 0 |
| GINGER1 (related TRs) | 0 | 0 | 0 | 0 | 6 | 0 |
| GINGER2 (related TRs) | 0 | 0 | 0 | 0 | 15 | 0 |
| CIN1 (related TRs) | 0 | 0 | 0 | 0 | 14 | 0 |
| Unclassified sequences | 7 | 5 | 2 | 0 | 2 | 21 |
| Total | 33 | 20 | 21 | 4 | 81 | 23 |
A. pisum is the first whole genome sequence of a basal hemimetabolous (exopterygota) insect(IAGC 2010) to be published. Shortly afterwards, another hemimetabolous genome was released (Kirkness et al. 2010), that of Pediculus humanus (human body louse), which has the smallest known insect genome, approximately four fold less than A. pisum (see Figure 2 below). With the exception of Rhodinus prolixus (for which the whole genome sequence is not yet available), the human body louse is the closest model insect to the pea aphid in the tree of life. However, a striking divergence in genomic retroelement content between them is apparent. P. humanus was reported to have only two full-length copies of LTR retrotransposons, both belonging to the Mdg1 clade of Ty3/Gypsy elements. A feasible explanation for this difference is the fact that the body louse is an obligate parasite restricted to humans, an extreme host specificity (not the case for A. pisum) that allows drastic genome reduction and, in the context of its exceptionally homogeneous environment, the P. humanus genome has “renounced” the possibility for adaptation offered by TEs in general and LTR retrotransposons in particular (González et al. 2008). Such a relative lack of retrotransposons compared with other related insects was also evident for the honey bee (Apis mellifera). In this case, an explanation based on the disruptive nature of retroelements in a genome exposed to selection every generation was hypothesized (HGSC 2006).
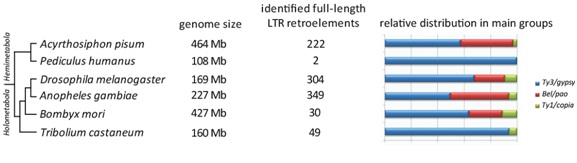
The number of LTR retroelements annotated in the A. pisum genome was not as high as expected from its genome size when compared with holometabolous (endopterygota) insects for which whole genome data are available (see Figure 2). For instance in Drosophila melanogaster (fruit fly), which has a genome less than half the size of the pea aphid genome, more than 300 full length LTR retrotransposons were identified (Kaminker et al. 2003). Similar retrotransposon numbers and a comparable genome size to that of D. melanogaster were reported for the dipteran insect Anopheles gambiae (Rho et al. 2008;Tubio et al. 2005). However, with a genome size comparable to that of A. pisum, in the lepidopteron Bombyx mori (silk worm) only 30 full-length retroelements have been identified (Rho et al. 2008). The genome of Tribolium castaneum (flour beetle) is comparable in size to that of dipterans yet it harbors a low number of retroelements (TGSC 2008), lower than that of the pea aphid. Assuming that draft genome analyses can vary depending on LTR element identification methodology and are being compared in forthcoming genomic assemblies (Sharakhova et al. 2006), general trends can be established. In this sense, the idea that the overall abundance of each type of transposable element correlates positively with genome size (Lynch & Conery 2003) does not apply here and no association between genome size and LTR retrotransposon content can be proposed among insects. Further investigation incorporating data from other insect genomes that have been released but without detailed information concerning LTR retroelements (HGSC 2006; Minervini et al. 2009; Nene et al. 2007; Werren et al. 2010) will help to lead to reliable conclusions. However, a general rule concerning the relative distribution of LTR retrotransposons among main groups can be inferred: Ty3/Gypsy-like elements are the most abundant in insect genomes.
