Element:TVCV
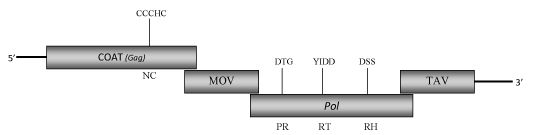
DescriptionTobacco vein clearing virus (TVCV) is a plant pararetrovirus detected only in the tobacco hybrid Nicotiana edwardsonii (N. clevelandii x N. glutinosa) where determines a typical foliar symptomatology, consisting in vein clearing to yellow mosaic and leaf deformation, from which derives its name Lockhart et al. 2000). TCVC belongs to Cavemovirus genus (for more details see ICTVdb) of Caulimoviridae plant virus family, and together to the Cassava vein mosaic virus (CsVMV) are the two species of Class 3 in the phylogenetic analysis performed by Llorens et al 2009. TVCV viral particles, of 50 nm in diameter, are composed of a 7767 bp dsDNA genome which each strand is interrupted by a site-specific discontinuity. The genome encodes for four open reading frames (ORFs) containing a COAT (gag) (ORF I) and pol-polyprotein (ORF III) with a Ty3/Gypsy-like domain organization (gag, protease, reverse transcriptase and RNaseH), a putative movement protein (MOV, ORF II) and a transcriptional activator region (TAV, ORF IV) typical of caulimoviruses. TVCV genes order, as well as that of CsVMV, is an unusual feature of these two pararetroviral species that differ from other Caulimoviridae genera in the position of their coat and movement regions, being the former upstream to the MOV-like gene (de Kochko et al. 1998; Calvert et al. 1995; Lockhart et al. 2000). The consensus RNA-binding sequence associated to the putative COAT-gag region, like that of other plant pararetrovirus, is larger than the related retroelement consensus RNA-binding sequence (C-X2-C-X4-H-X4-C) because it includes an additional cysteine (Cys) at the -2 position (C-X-C-X2-C-X4-H-X4-C) (Hull 1996; Bouhida et al. 1993). TVCV apparently is transmitted only vertically (by seed to 100% of progeny plants) because of the experimentally transmission tentative from N. edwardonii to any other Nicotiana spp. or other plant species by mechanical inoculation, grafting or insect (aphid) vectors, failed (Lockhart et al. 2000; Harper et al. 2002). This evidence suggests that TVCV, like the other two integrated pararetrovirus Banana streak virus (BSV) and Petunia vein clearing virus (PVCV), could exist as an integrant sequence in the host genome and that under certain conditions (i.e. response to stress) becomes episomally active, causing the disease development (Harper et al. 2002). No TVCV integrants have been found in the genome of the female parent N. clevelandii while the male parent N. glutinosa, although contains integrants in numerous loci of its genome, never develops disease symptoms or episomal TVCV infection (Harper et al. 2002). Interestingly, the nucleotide sequence and genome organization of TVCV show high similarities to those of a pararetrovirus-like sequences (TPV) described in high copy number in the genome of N. tabacum (Jakowitsch et al. 1999). Altogether these data suggest that the episomal form of TVCV probably arises from integrated sequences present in the genome of N. edwardsonii inherited from the male parent N. glutinosa and is related to pararetroviral-like elements occurring in other Nicotiana spp. (Lockhart et al. 2000). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||