Element:SbCMV
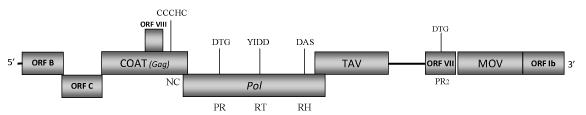
DescriptionSoybean chlorotic mottle virus (SbCMV) is a plant pararetrovirus isolated from soybean (Glycine max) plants (Iwaki et al. 1984). It is the type species of the genus Soymovirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). SbCMV is easily transmitted by mechanical inoculation, while attempts to transmit it by seed or insect vectors have failed (Iwaki et al. 1984). Thus, the biological vector of this virus remains unknown. SbCMV provokes mosaic and stunting symptoms in naturally infected plants whereas on mechanically inoculated soybean plants could determine a different symptomatology depending on the cultivar (i.e veinclearing, chlorosis and reduced size of young leaves, as well as mottling and leaf roll, Iwaki et al. 1984; Takemoto and Hibi 2001). Morphologically, SbCMV has spherical virions of approximately 50 nm in diameter (Iwaki et al. 1984) containing a 8178 bp double-stranded DNA genome. This genome shows three single-stranded discontinuities which are typical of caulimoviruses: one (G1) in the (-) strand and two (G2 and G3) in the other (+) strand (Hasegawa et al. 1989); as well as eight/nine open reading frames (ORFs) designated as Ia, Ib, II, III, IV, V, VI, VII, VIII (Hasegawa et al. 1989; Takemoto and Hibi 2001; 2005). ORF I splits into two smaller ORFs, Ia and Ib, which are separated by a single in-frame stop codon. ORF Ia encodes a putative protein of 303 amino acids (aa) associated to cell-to-cell movement (MOV), while ORF Ib encodes a 103 aa long protein product whose function is so far unknown. This ORF also includes a putative tRNAMet primer-binding site (PBS), which has been demonstrated to be essential for virus replication (Takemoto and Hibi 2001). ORF II (namely here as ORF B) encodes a protein of 201 aa also preserved in other soymoviruses whose function is still unclear (Takemoto and Hibi 2001) but it has been demonstrated that it is required for virus replication or assembly (Takemoto and Hibi 2001; 2005). The product of ORF III (namely here as ORF C) contains a putative coiled-coil domain in its N-terminal region that has been found to be indispensable for virus infection (Takemoto and Hibi 2001). Our sequence analyses show that ORFC-product reveals similarities with the related ORFs C of the other soymoviruses (BRRV and CmYLCV) and with the ORF III (gp3) of Caulimovirus RuFDV. The function of these ORFs has not been yet well defined and their relationship with the "virion associated protein" (VAP) of the other Caulimoviridae species remains to be demonstrated. ORF IV codes for the gag-like COAT, one of the more conserved genes among caulimoviruses and retroelements (Hull 1996; Bouhida et al. 1993). ORF V codes for a pol polyprotein of 692 aa displaying the typical aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. ORF VI encodes a putative TAV protein of 462 aa showing the presence of a translational transactivator motifs (Hasegawa et al. 1989; Takemoto and Hibi 2001). ORF VII potentially encodes a protein of 148 aa whose function remains unknown although our analyses reveal the existence of a putative second protease-like domain which aa sequence shows similarity to that of the conventional Pol-protease. We have also identified the presence of a similar domain in the corresponding ORFs VII of BBRV and PCSV soymoviruses, as well as in the genome of the tungrovirus Rice tungro bacilliform virus (RTBV) and of the cavemovirus Cassava vein mosaic virus (CsVMV), but it is not clear if these sequences are functionally active proteases additional to those found in the pol genes. An eighth small ORF (ORF VIII) is located within the gag-like gene but its function remains to be still demonstrated (Hasegawa et al. 1989). Between ORFs VI and VII is present a large intergenic region of approximately 500 nucleotides that contains a putative promoter region (promoter IV) which expression activity has been demonstrated to be similar to that of the CaMV 35S promoter, one of the strongest known promoters active in plants (Hasegawa et al. 1989; Conci et al. 1993). StructureFigure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||