Element:SIRE1-4
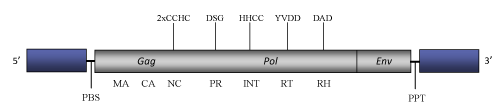
DescriptionSIRE1 is a Ty1/Copia LTR retroelement described in the genome of Glycine max (Laten 1999). SIRE1 belongs to SIRE a clade of Ty1/Copia plant LTR retrotransposons and potential retroviruses (Fauquet et al. 2005) evolutionarily related to another clade of plant Ty1/Copia LTR retrotransposons called Oryco (Llorens et al. 2009). SIRE clade gives the name to a genus within the current ICTV classification of the Ty1/Copia into three genera – Pseudovirus, Hemivirus and Sirevirus (Boeke et al. 2005). The genome structure of SIRE1-4 (taken as a SIRE1 representative) is 9.8 Kb in size (9805 bp in length) including two long LTRs of 1148 bp (Laten et al. 2003). The internal region of this element contains a Primer Binding Site (PBS), complementary to a tRNAiMet, two Open Reading Frames (ORFs) and a Polypurine Tract (PPT). The largest first ORF contains a gag-pol gene encoding for the tyìcal gag and pol protein domains (gag, protease, integrase, reverse transcriptase and RNase H). This ORF is followed by a second ORF separated by a single stop codon that encodes for a putative env protein (Laten et al. 2003). In comparison to other Ty1/Copia elements, a taxonomically relevant feature specific of Sire-like elements is their extremely large gag c-terminus consistent with the NC domain, which may display two or three CCHC arrays (Llorens et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||