Element:HTLV-1
DescriptionHuman T-Cell Leukemia Virus (HTLV-1) is a complex retrovirus responsible of inducing adult T-cell leukemia (ATL) in humans (Poiesz et al. 1980). HTLV-1 is associated with a neurologic degenerative disorder known as tropical spastic paraparesis (TSP) or more commonly, HTLV-1-associated myelopathy (HAM). Currently, there are 20 million people worldwide infected with HTLV-1 (Edwards et al. 2003), which is spread via semen (sex), needles, milk, and blood. Several evidences suggest that HTLV-1 may have arisen from interspecies transmission between STLV-1-infected monkeys and humans, followed by evolution in the human host (Masahiro et al. 1996). Phylogenetic analyses have revealed four HTLV-1 molecular subtypes:
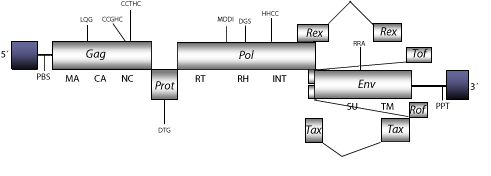
The genomic structure of HTLV-1 is 8.5 Kb in size including LTRs of 986 nt (pro-5´LTRs and pro-3´LTR of 405-581 nt, respectively). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNAPro, Open Reading Frames (ORFs) for gag, pol, and env genes. HTLV-1 displays a region termed the pX region, which contains the accessory genes rex and tax (both essential for viral replication and cellular transformation in infected cells), as well as two ORFs that encode for the proteins Tof and Rof (see accessory genes), and the Polypurine Tract (PPT) common to all retroviruses (Malik, Even and Karpas 1988; Yoshida et al. 1995; Koralnik et al. 1992; Pique et al. 2000). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||