Element:Gmr1
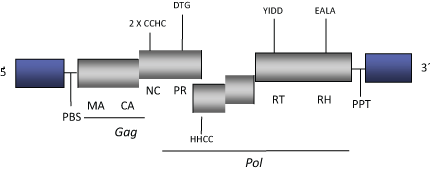
DescriptionGmr1 is a unsual Ty3/Gypsy LTR retrotransposon described (between two immunoglobulin light-chain L1 regions) in the genome of the Atlantic cod Gadus morhua, together with other elements from diverse vertebrates, including the zebrafish Danio rerio and Xenopus (Butler et al. 2001; Goodwin and Poulter 2002). These constitute an own clade (to which Gmr1 gives the name) possibly related via sequence similarity with Osvaldo clade (Butler et al. 2001; Goodwin and Poulter 2002; Llorens et al. 2009). Interestingly, Gmr1 elements differ from other Ty3/Gypsy LTR retroelements in the fact that the integrase domain lies upstream of the reverse transcriptase domain, an arrangement which is characteristic of Ty1/Copia LTR retroelements (Butler et al. 2001; Goodwin and Poulter 2002). The genomic structure of Gmr1 is 8 Kb in size including LTRs of approximately 1 Kb in size. The internal region of this element displays a Primer Binding Site (PBS) complementary to a tRNAala, an Open Reading Frame (ORF) for gag and protease displaying a frameshifting and overlapping with the integrase ORF which shows a frameshifting and another ORF for reverse transtriptase and RNAseH. The element shows the typical Polypurine Tract (PPT) adjacent to the 3´LTRs (Butler et al. 2001;Goodwin and Poulter 2002). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||