Element:FIV
DescriptionFeline Immunodeficiency Virus (FIV), also called Feline T-lymphotropic Virus (FTLV), is a complex retrovirus similar to HIV. This retrovirus was originally isolated from infected cats in peripheral blood lymphocytes (Pedersen et al. 1987). Due to its induction for an immunodeficiency status in domestic cats, this retrovirus was later called FIV (Yamamoto et al. 1988a). Along with Simian Immunodeficiency Viruses (SIVs), FIV is considered to be an important AIDS infection model in small animals for helping to develop therapy against the HIV infection (Hohdatsu et al. 2000). FIV is usually inoculated through infected cat bites (Pedersen et al. 1989) and several studies suggest that male street cats are the main group of risk (Yamamoto et al. 1988b; Grindem et al. 1989). The reversion of the blood ratio between CD4+ and CD8+ lymphocytes has been reported by several authors in infected and symptomatic cats (Novotney et al. 1990; Tompkins et al. 1991; Hoffmann-Fezer et al. 1992). The infection presents three stages of clinic symptomatology:
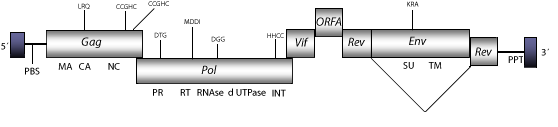
The genomic structure of FIV is 9.4 Kb in size including LTRs of 360 nt. The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNALys3, Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, orf1 (vif) and rev (both expanded in two exons), which are two accessory genes common in other lentiviruses, orf2 (A), and adjacent to the 3´LTR the Polypurine Tract (PPT) common to other LTR retroelements (Talbott et al. 1989; Phillips et al. 1990). As other non-primate lentiviruses, FIV displays between the RNase H and INT domains, an ORF coding for a dUTPase similar to that observed in betaretroviruses, N-terminal to the PR domain (Elder et al. 1992; Payne and Elder 2001 and references therein). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||