Element:EFV
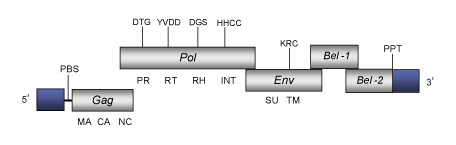
DescriptionEquine foamy virus (EFV) is a foamy retrovirus originally isolated from equine species. EFV belongs to genus Spumaretrovirus within Retroviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. (2009) is located within Class 3 of the Retroviridae family. Similar to other foamy virus, EFV exhibits a highly characteristic ultrastructure and induces syncytium formation and subsequent cell lysis on a large number of cell lines (Tobaly-Tapiero et al. 2000). The genomic structure of EFV is 12.035 Kb in size including LTRs of 1.449 bp (Tobaly-Tapiero et al. 2000). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNALys, three Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, two 3´accessory genes, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Tobaly-Tapiero et al. 2000). The predicted Gag protein of EFV consists of 559 amino acids; the ORF of the pol gene -which harbors the protease, RT, RNase H and integrase domains- begins at position 3157 and consists of 1154 residues. The env gene encodes a predicted 987-residue and downstream to this ORF are located the two accessory genes, Tas-1 (ORF 1) and Bel-2 (Tobaly-Tapiero et al. 2000). The first one, the Tas-1 ORF (the spumaretroviruses transactivator, also called Bel 1) transactivates viral gene expression by direct binding to the viral DNA on specific sequences in the long terminal repeat (LTR) and in an internal promoter (IP) (Tobaly-Tapiero et al. 2000; Lecellier et al. 2002). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||