Element:CYMV
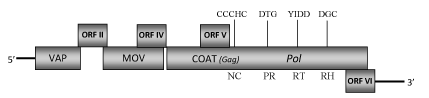
DescriptionThe plant pararetrovirus Citrus yellow mosaic virus (CYMV), also called Citrus yellow mosaic badnavirus (CMBV), is the aetiological agent of "Citrus mosaic disease", a disorder first described in India in 1970s (Dakshinamurti and Reddy 1975) and actually widely distributed as a common and severe disease, especially in sweet orange (Ahlawat et al. 1984, 1996a,b). The virus has been reported from four Citrus species: Sweet orange (Citrus sinensis, AF347695), Rangpur lime (C. limonia, EU884191), Acid lime (C. aurantifolia, EU489744) and Pummelo (C. grandis, EU489745) (Huang and Hurtung 2001; Borah et al. 2009). The sequence corresponding to the Rangpur lime CYMV-isolate is that used as reference sequence in our analyses. CYMV is a member of Caulimoviridae family genus Badnavirus (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 2 of the Caulimoviridae family. CYMV has a circular double-stranded DNA genome of about 7.5 Kb in size contained in bacilliform-shaped particles of approximately 30x130 nm. It encodes six-seven Open Reading Frames (ORFs) arranged in only one sense and containing the domains highly conserved among all plant pararetroviruses (Huang and Hurtung 2001; Borah et al. 2009). The putative ORF I protein shows sequence similarity to the virion associated proteins (VAPs) encoded by the corresponding Caulimoviridae-ORFs I. ORF II potentially encodes a protein of 15.5 kDa in which have been identified two conserved domains (KQNN and KDPY) homologous to those of the Cacao swollen shoot virus (CSSV)-ORF II product that seems to possess a DNA-binding property (Borah et al. 2009; Jacquot et al. 1996). In contrast to other isolates, CYMV from Rangpur lime (a species used as rootstock in citrus propagation) is the only one containing seven ORFs because its ORF III is split into two parts (ORF IIIa and ORF IIIb) due to the presence of three stop codons downstream the movement protein region (MOV). In addition to the two cysteine-rich tracts "C-X-C-X2-C-X4-H-X4-C" and "C-X2-C-X11-C-X2-C-X4-C-X2-C" associated to the gag-like region- only observed in Badna- and Tungroviruses and similar to those identified in the nucleocapsid domains of LTR retroelements (Hull 1996; Bouhida et al. 1993; Llorens et al. 2009)- ORF III also contains domains homologous to those of the aspartic protease (PR), the reverse transcriptase (RT) and RNase H (RH) of all other known Badnavirus species (Borah et al. 2009). Other small additional ORFs displayed by CYMV genome potentially encode for proteins with a molecular weight of 17.4 kDa (ORF IV), 10.7 kDa (ORF V) and 17.5 kDa (ORF VI). Additional ORFs resembling to CYMV-ORF IV and similarly located within their respective ORF III have also been identified in the genome of other badnaviruses (Su et al. 2007; Yang et al. 2003). The CYMV-ORF V product has limited similarity to those of ORFs IV and V of Cassava vein mosaic virus (CsVMV), as well as to the Soybean chlorotic mottle-like virus (SbCMV)-ORF VIII, while the predicted product of ORF VI shows some homologies with the putative product of CSSV-ORF Y (Huang and Hurtung 2001). Although these proteins show no significant similarities to those in the databases, conservation of these additional ORFs among the four CYMV isolates would suggest they are functional and are likely expressed (Borah et al. 2009). CYMV is transmitted by grafting, by its natural vector Planococcus citri as well as by Agroinoculation with Agrobacterium tumefaciens transformants (Huang and Hurtung 2001; Huang and Hurtung 2008). The infected plants show the characteristic "Citrus mosaic disease" symptomatology consisting in yellow mosaic of the leaves and yellow flecking along the veins (Ahlawat et al. 1996a). The affected trees suffer significant severe losses in production and in fruit quality (Ahlawat et al. 1996a). StructureFigure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||