Element:CERV
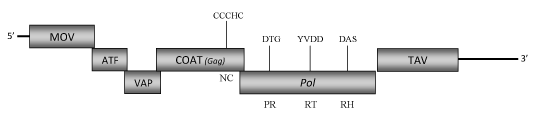
DescriptionCarnation etched ring virus (CERV) is a plant pararetrovirus species of the genus Caulimovirus (Caulimoviridae family, International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) that causes the "Etched ring disease" on carnation plants (Dianthus caryophyllus) characterized by necrotic flecks, rings and line patterns on the leaves. The CERV Indian isolate has been included in our phylogenetic analyses (see, Llorens et al 2009). CERV isometric virions are composed of a genomic circular double-stranded DNA (7924 bp long) encoding for six Open reading frames (ORFs) whose size and organization are similar to those of known caulimoviruses (Raikhy et al. 2005, unpublished; Hull et al. 1986). The aminoacids sequences of the protein products show similarities with those encoded by the corresponding ORFs of known Caulimoviruses. ORF I encodes the movement protein (MOV), ORF II and ORF III are similar to the "aphid transmission factor" (ATF) and "virion associated protein" (VAP), respectively. ORF IV encodes the COAT protein and contains the "RNA-binding" motif characteristic of the Caulimoviridae Gag-like domain (C-X-C-X2-C-X4-H-X4-C ) (Bouhida et al. 1993;Hull 1996; Llorens et al 2009). ORF V product is a polyprotein displaying the typical Pol domains -aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) – and finally ORF VI encodes for a "Translational transactivator protein" (TAV) (Raikhy et al. 2005, unpublished). Between ORFs V (Pol) and VI (TAV) the virus presents a short intergenic region like those observed in the genome of other Caulimovirus species such as EVCV and LLDAV. A second larger intergenic region is downstream to the ORF VI. A promoter, located in the 3′ terminus of ORF VI and extending into the large intergenic region of CERV (Kuluev and Chemeris 2007), has been demonstrated to display an expression activity similar or more efficient in some plants than that of the CaMV 35S promoter (Kuluev et al. 2008). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||