Element:BSMyV
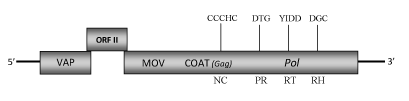
DescriptionBanana streak virus (BSV), the causal agent of viral "leaf streak disease" of banana and plantain (Musa spp.) (Lockhart, 1986), is a member of the plant pararetrovirus genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). Several isolated have been studied, one of which is BSMyV infecting a triploid Musa cultivar "Mysore’" (AAB group), an Indian hybrid originating from Musa acuminata (A genome) and Musa balbisiana (B genome) species (Geering et al. 2005a). According to Llorens et al. 2009, BSMyV belongs to Badnavirus genus within Class 2 of the Caulimoviridae family. The BSMyV has bacilliform particles containing a circular double-stranded DNA genome of about 7.6 kb in size (7650 bp long) that encodes for three Open reading frames (ORFs I, II, III) (Geering et al. 2005a). Unlike other species of the genus, the beginning of ORF I does not start with a conventional AUG start codon, however it shows the same sequence similarity to the caulimoviruses-related virion associated protein (VAP). ORF II encodes for an undescribed protein with a molecular mass of 14.6 kDa, while the ORF III displays the polyprotein domains characteristic of this genus [MOV, COAT(gag), PR, RT, RH] (Geering et al. 2005a). The gag-like region includes a large region (only observed in Badna- and Tungroviruses), which is rich in zinc finger (CCHC) array duplications similarly to those of several LTR retroelement nucleocapsids (Hull 1996; Bouhida et al. 1993; Llorens et al. 2009). The viral "leaf streak disease", first described in Ivory Coast, Africa in 1968 (Lassoudiere 1974), now is probably distributed worldwide in cultivated banana and plantain crops. Its symptomatology is correlated with variations in virus concentration. The symptoms may vary from faint broken chlorotic lines to necrosis of young leaves, internal necrosis of the pseudostem and plant death. The bunch size and fruit quality are also compromised (Lockhart, 1986). The virus is transmitted naturally by the two mealybug vectors (Pseudococcidae) Planococcus citri and Saccharicoccus sacchari (both of which colonize banana), as well as by seed, even if the vegetative propagation is the principal method of virus spread. The ability of P. citri to act as a vector of BSMyV has been demonstrated (Geering et al. 2005a). Althouth BSV genome does not contain any integrase motif, multiple DNA sequences related to the virus genome are found integrated into the nuclear genome of different Musa cvv. (Harper et al. 1999; Ndowora et al. 1999; Geering et al. 2005b; Gayral et al. 2008; Gayral and Iskra-Caruana 2009). Two types of BSV-Endogenous pararetrovirus (EPRV) sequences have been described in banana. The first type sequences, defined as noninfectious (with nonfunctional viral ORFs and/or incomplete viral genomes), are present in the two most common Musa species Musa acuminata (denoted A) and Musa balbisiana (denoted B) that, together to Musa schizocarpa (denoted S) are the wild progenitors of the domesticated banana (Geering et al. 2005b). The second EPRV sequences, so-called infectious type, contain the complete functional viral genome and have been detected in some Musa genotypes (i.e AAB and BB groups, Geering et al. 2005b ; Gayral et al. 2008). Geering et al. (2005a) suggest that BSMyV-DNA is also integrated in the B genome of Musa but it is still unclear whether BSMyV-EPRV is able arise a functional episomal genome. Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||