Ty3/Gypsy
Introduction:
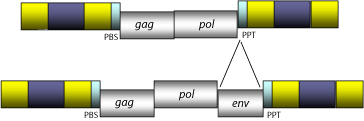
Ty3/Gypsy LTR retroelements constitute a family of retroviruses and LTR retrotransposons widely distributed in plants, fungi and animals. They are central to understand the evolutionary history of the LTR retroelement system as they show, depending on the lineage, relationships in both sequence similarity and genomic structure with the remaining families of LTR retroelements (for more information see Llorens et al. 2009). Of particular interest is the common evolutionarily history between several Ty3/Gypsy lineages and Retroviridae retroviruses (see, Eickbush and Malik 2002; Llorens et al. 2008). In fact, the main difference between a retrovirus and a LTR retrotransposon is that the retrovirus has an additional Open Reading Frame (ORF) coding for an envelope (env) polyprotein necessary for transferring retroviruses cell-to-cell (see Retroviridae family). Upon this, it is now known that env-like genes are not exclusive of Retroviridae, as several lineages belonging to not only Ty3/Gypsy but also other LTR retroelement families have been shown to also be carriers of env genes (see Eickbush and Malik 2002). Particularly, many studies demonstrate that the Gypsy element of D. melanogaster is capable to act as a retrovirus and horizontally transfer itself from a Drosophila species to another (Mizrokhi and Mazo 1991; Kim et al. 1994; Song et al. 1994; Alberola and de Frutos 1996; Terzian et al. 2000). Although the possibility that functional retroviruses might also exist in plants is still unclear, the chance by which any LTR retrotransposon could become a potential retrovirus when acquiring an env gene is fascinating issue of discussion (Kumar 1998 ; Kumar and Bennetzen 1999; Zaki 2003).
Genomic structure:
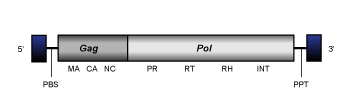
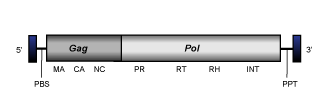
Similarly to vertebrate endogenous retroviruses (see Retroviridae family), a canonical Ty3/Gypsy LTR retroelement (in their inserted genomic state) usually show a genome of variable size (between 4 and ~15Kb) flanked by LTRs and structured as follows:
- A 5' long terminal repeat (LTR) of 100-2000 nt
- A non-coding region of 75-250 nt that corresponds to the first portion of the retrotranscribed genome.
- A Primer Binding Site (PBS) of 18 nt, complementary to a specific zone of the 3' end of a tRNA normally provided by the host cell in order to start the retrotranscription.
- Open Reading Frames (ORFs) for gag, pol, (and env in retroviruses) genes and other accessory genes (in the case of several lineages such as Tat clade and diverse chromoviruses).
- A small region of ~10 A/G "Polypurine Tract" (PPT), responsible for starting the synthesis of the proviral (+) DNA strand.
- A 3' long terminal repeat (LTR) of 100-2000 nt
Evolutionary history and in-progress taxonomy:
The phylogenetic analysis of Ty3/Gypsy LTR retroelements inferred based on the gag-pol common internal region suggests that all protein domains coded by these elements follow a unique evolutionary history originally reported from RT analyses (Xiong and Eickbush 1990). In a step forward, Ty3/Gypsy LTR retroelements were originally classified into two major genera called Metaviridae and Errantiviridae (for more information, see (Eickbush and Jamburuthugoda 2008). For some time, this classification has been an important reference in LTR retroelement taxonomy, but it is now considered inconclusive by many authors because sequencing projects have revealed new lineages, updating the original perspective. According to Llorens et al. 2009, the inferred pol phylogeny of Ty3/Gypsy LTR retroelements inferred based on pol (PR-RT-RNaseH-INT) reveals two large branches. This is additionally supported by phylogenetic reconstruction analysis of this family inferred based on both gag and pol polyproteins (see Ty3/Gypsy gag-pol tree). The first branch encompasses all chromodomain-INT-containing LTR retrotransposons, usually called chromoviruses (Marin and Llorens 2000), which includes at least two well-supported clusters, in this database named as "Plants" and "Fungi/Vertebrates". The second Ty3/Gypsy branch referred to as Branch 2, encompasses the remaining lineages of non-chromoviral Ty3/Gypsy LTR retrotransposons and retroviruses of plants and animals.
| Host phyla | Branch | Genus and/or cluster | clade | Env | Chromodomain |
|---|---|---|---|---|---|
| Viridiplantae | Branch 1 (Chromoviridae) | Plants | CRM | no | no |
| Viridiplantae | Branch 1 (Chromoviridae) | Plants | Del | no | yes |
| Viridiplantae | Branch 1 (Chromoviridae) | Plants | Galadriel | no | yes |
| Viridiplantae | Branch 1 (Chromoviridae) | Plants | Reina | no | yes |
| Chlorophyta | Branch 1 (Chromoviridae) | Plants | REM1 | no | yes |
| Cryptophyta | Branch 1 (Chromoviridae) | Plants | G-Rhodo | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | Pyggy | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | MGLR3 | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | Pyret | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | Maggy | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | MarY1 | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Undefined | Tse3 | no | yes |
| Fungi | Branch 1 (Chromoviridae) | Fungi and vertebrates | TF1-2 | no | no |
| Fungi | Branch 1 (Chromoviridae) | Undefined | Ty3 | no | no |
| Vertebrata | Branch 1 (Chromoviridae) | Fungi and vertebrates | V-clade | no | yes |
| Amoebozoa | Branch 1 (Chromoviridae) | Undefined | Skipper | no | yes |
| Viridiplantae | Branch 2 | Athila/Tat | Athila | yes | no |
| Viridiplantae | Branch 2 | Athila/Tat | Tat | no | no |
| Arthropoda | Branch 2 | Errantiviridae | 17.6 | yes | no |
| Arthropoda | Branch 2 | Errantiviridae | Gypsy | yes | no |
| Arthropoda | Branch 2 | Metaviridae | 412/mdg1 | no | no |
| Arthropoda | Branch 2 | Metaviridae | Micropia/mdg3 | no | no |
| Bilateria | Branch 2 | Mag | A-clade | no | no |
| Bilateria | Branch 2 | Mag | B-clade | no | no |
| Bilateria | Branch 2 | Mag | C-clade | no | no |
| Deuterostomia | Branch 2 | Metaviridae | Gmr1 | no | no |
| Arthropoda | Branch 2 | Metaviridae | Osvaldo | Only Osvaldo | no |
| Nematoda | Branch 2 | Metaviridae | Cer2-3 | no | no |
| Nematoda | Branch 2 | Metaviridae | Cer1 | no | no |
| Protostomia | Branch 2 | Metaviridae | CsRN1 | no | no |
| Deuterostomia | Branch 2 | Metaviridae | Tor1 | no | no |
| Deuterostomia | Branch 2 | Metaviridae | Tor4 | some elements | no |
| Deuterostomia | Branch 2 | Metaviridae | Tor2 | no | no |
| Deuterostomia | Branch 2 | Metaviridae | Cigr-1 | no | no |
Branch 1 (Chromoviruses)
Chromoviruses, also called Class B (Wright and Voytas 1998) or Ty3 group (Malik and Eickbush 1999), constitute the major branch of Ty3/Gypsy lineages described in genomes of plant, fungi, and vertebrate organisms. Chromoviruses usually bear a chromodomain at the C-terminal end of their integrases (Malik and Eickbush 1999). From this evidence, the term "Chromoviridae" (Marin and Llorens 2000) has been proposed to describe this branch within the Ty3/Gypsy phylogeny. The chromoviral diversity has been found to be greater than previously supposed and their origins have been significantly extended by the finding of representatives in the most basal plant lineages, those of green and red algae (Gorinsek, Gubensek and Kordis 2004; 2005). This suggests that this group indeed represents the oldest and most widespread group of Ty3/Gypsy elements (see also Kordis 2005). Chromoviral species at present contemplated in this database may be divided in two well-supported major clusters, for simplicity´s sake named Plants and Fungi/Vertebrates and several lineages with unclear classification.
Plants
This cluster collects chromoviral elements described not only in Archaeplastida organisms (green plants) but also in several algae protists. Plant chromoviruses normally use a methionine starting tRNA (iMet) (Levin 1995 ; Butler et al. 2001) and split into at least six clades as follows (note however that sequencing project continuously reveal new lineages).
CRM
CRM elements are plant centromere-specific Ty3/Gypsy LTR retrotransposons whose integrases exhibit a chromodomain region substantially divergent from the overall chromodomain consensus (Gorinsek, Gubensek and Kordis 2004). CRM elements usually have an internal region of 6-7 Kb in size, flanked by LTRs of 0.5-1 Kb and characterized by the presence of a Primer Bindind Site (PBS), two ORFs for the canonical gag and pol genes typically observed in LTR retrotransposons, and a Polypurine Tract (PPT) adjacent to the 3´LTR.
Galadriel
Galadriel elements constitute a clade of plant Ty3/Gypsy LTR retrotransposons (Gorinsek, Gubensek and Kordis 2004) characterized by an internal region of 5.2-6 Kb in size, flanked by LTRs of 0.4-0.8 Kb and by the presence of a PBS, two ORFs for the gag and pol genes characteristic of canonical LTR retrotransposons, and a PPT adjacent to the 3´LTR. All Galadriel elements code for chromodomain-integrases.
Reina
Reina elements are plant Ty3/Gypsy LTR retrotransposons (Gorinsek, Gubensek and Kordis 2004) displaying a genome of 4.7-5 Kb in size, flanked by LTRs of 0.35 Kb and characterized by an internal region that presents a PBS, the gag and pol genes typically observed in LTR retrotransposons, and a PPT adjacent to the 3´LTR. All Reina elements codify for chromodomain-integrases.
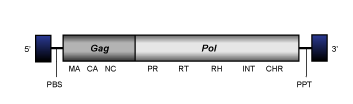
Del
Del clade (Llorens et al. 2008) also and previously described as Tekay clade (Gorinsek, Gubensek and Kordis 2004) constitutes a plant Ty3/Gypsy LTR retrotransposons, which have an internal region of 6.2-10.2 Kb in size, flanked by LTRs of 1.1-4.4 Kb and characterized by the presence of a PBS, two ORFs for the gag and pol genes, which are common in almost all LTR retrotransposons, and a PPT adjacent to the 3´LTR. All Del elements codify for chromodomain-integrases.
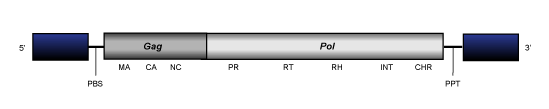
REM
REM is a single-element clade representing REM1, a LTR retrotransposon described in the genome of the green algae Chlamydomonas reinhardtii (Perez Alegre et al. 2005). Although REM1 shows the typical gag-pol genome organization of Ty3/Gypsy chromoviruses it present a reorganization and partial lost of the INT domain, which is genomically located in reverse frame and upstream to gag probably due to an ancient rearragement of this element during evolution (Llorens et al. 2009) (for more details see the file of REM1 element).
G-Rhodo
G-Rhodo (Gypsy Rhodomonas) is representative of a clade of chromoviral Ty3/Gypsy LTR retrotransposons to date constituted by a single element and described, together with another Ty1/Copia element, in the unicellular eukaryotic algae Rhodomonas salina (Khan et al. 2007) (for more details see the file of G-Rhodo element).
Fungi and vertebrates
The cluster of fungi and vertebrate chromoviruses collects chromoviral elements described in the genomes of both fungi and vertebrate organisms (only in fishes and amphibians as full-lenght retrotransposons). Interestingly, the PBS of fungi and vertebrate chromoviruses differs from that used by plant chromoviruses: while plant chromoviruses use a methionine starting tRNA (iMet), fungi and vertebrate chromoviruses use their own self-priming mechanism to start the reverse transcription (Levin 1995 ; Butler et al. 2001). This cluster splits into at least seven clades as detailed below (although sequencing projects constantly reveal new lineages).
TF1-2
TF1-2 consists of a Schizosaccharomyces pombe clade collecting two of the most widely studied LTR retrotransposons closely related each other - TF1 and TF2 (Levin, Weaver and Boeke 1990; Weaber et al. 1993). The internal region of these two elements is 4.4-4.9 Kb in size, is flanked by LTRs of 0.35 Kb and presents a PBS, ORFs for the gag and pol genes, and a PPT adjacent to the 3´LTR. Although both phylogenetic and comparative analyses reveal that these two elements belong to the chromoviral branch of Ty3/Gypsy LTR retrotransposons, their encoded Integrase products do not carry any recognizable chromodomain (a typical feature of chromoviruses).
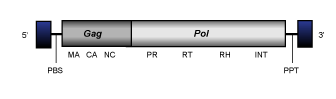
Pyggy
Pyggy elements constitute a clade of Pezizomycotina Ty3/Gypsy LTR retrotransposons (Llorens et al. 2009) of 5.2-6 Kb in size and usually flanked by LTRs of aproximately 0.2 Kb. The internal region displays PBS, two ORFs (usually displayed in different frames) for the gag and pol genes characteristic of canonical LTR retrotransposons, and a PPT adjacent to the 3´LTR. All Pyggy elements code for chromodomain-integrases.
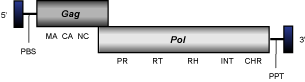
MGLR3
MGLR3 is a chromodomain-integrase encoding LTR retrotransposon characterized in the genome of Magnaporthe grisea (Kang 2001) which represents a clade within fungal chromoviruses (Gorinsek, Gubensek and Kordis 2004). In this GyDB iteration, MGLR3 represents a single-sequence clade (see the file of MGLR3 element), but it has other relatives reported, which monophyletically group MGLR3 and Pyggy clades in a single cluster (for more details, see Gorinsek, Gubensek and Kordis 2004).
Maggy
Maggy elements constitute a clade of Ascomycota Ty3/Gypsy LTR retrotransposons (Gorinsek, Gubensek and Kordis 2004) of 5-8 Kb in size and usually flanked by LTRs of aproximately 0.25 Kb. The internal region of these elements displays PBS, the different gag and pol genes characteristic of canonical LTR retrotransposons organized in two ORFs and a PPT adjacent to the 3´LTR. All Maggy elements code for chromodomain-integrases.
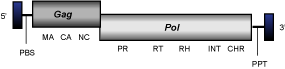
MarY1
MarY1 is a chromodomain-integrase encoding LTR retrotransposon characterized in the genome of Tricholoma matsutake (Murata and Yamada 2000), which represents a clade within fungal chromoviruses (Gorinsek, Gubensek and Kordis 2004). In this GyDB iteration, marY1 represents a single-sequence clade (see the file of marY1 element) but has other relatives reported (for more details, see Gorinsek, Gubensek and Kordis 2004).
Pyret
Pyret elements constitute a clade of Ascomycota Ty3/Gypsy LTR retrotransposons (Gorinsek, Gubensek and Kordis 2004) of 7-8 Kb in size and usually flanked by LTRs of aproximately 0.4-05 Kb. The internal region of these elements displays PBS, the different gag and pol genes characteristic of canonical LTR retrotransposons organized in one, two or three ORFs (depending on the retroelement species) and a PPT adjacent to the 3´LTR. All Pyret elements code for chromodomain-integrases.
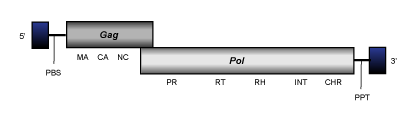
V-clade
V-clade (Llorens et al. 2009) covers the known to date diverse and active chromoviruses of fishes and amphibians and related to Amn-ichi (Piskurek et al. 2008) and Sushi-ichi (Poulter and Butler 1998). In consensus, these elements are 5 Kb in size, including LTRs of 0.2-0.5 Kb, they present PBS, the typical gag and pol genes (arranged in one or two ORFs), and a PPT adjacent to the 3´LTR. All V-clade elements described to date show chromodomains at the C-terminus of their integrases.
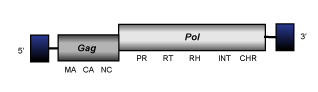
Undefined clades
The chromoviral branch includes diverse elements, which have unclear classification as they show a certain degree of divergence from other chromoviruses. Three examples are provided below. Although phylogenetic analyses may (in several cases) collect these three in a single clade, comparative analyses do not reveal strong similarity among them. This suggests that instead of a direct ancestor, the common feature among these three elements is that they are in the Felsestein zone of the chromoviruses tree (they group altogether because they are the most distant elements within the chromoviruses radiation).
Ty3
Ty3 is a LTR retrotransposon with specificity for integrating next to tRNA genes in Saccharomyces cerevisiae (Clark et al. 1988; Hansen, Chalker, and Sandmeyer 1988; Hansen and Sandmeyer 1990). It constitutes a small clade within chromoviruses in which two Ty3 subtypes were characterized (Hansen, Chalker, and Sandmeyer 1988). Ty3 encodes for the typical gag and pol ORFs and although Ty3 belongs to the chromoviral branch of Ty3/Gypsy LTR retrotransposons it usually falls outside of the fungi/vertebrate cluster in phylogenetic analyses. The integrase of this element lacks the chromodomain characteristic of chromoviruses. Moreover, the priming mechanism of this element differs from that other fungal and vertebrate chromoviruses as it displays, similarly to plant chromoviruses, a primer binding site (PBS) complementary to a tRNAiMet (for more details see the file of Ty3 element).
Skipper
Skipper is a chromodomain-integrase encoding LTR retrotransposon characterized in the genome of Dictyostelium discoideum (Leng et al. 1998) and represents a single-sequence clade within chromoviruses (for more details see the file of Skipper element).
Tse3
Tse3 is a LTR retrotransposon characterized in the genome of Saccharomyces exiguus (Neuveglise et al. 2002) and represents a single-sequence clade within chromoviruses. Tse3 encodes for the typical gag and pol ORFs, and belongs to the chromoviral branch of Ty3/Gypsy LTR retrotransposons but falls outside of the fungi/vertebrate cluster in phylogenetic analyses. The integrase encoded by Tse3 lacks the chromodomain typically observed in chromoviral integrases (for more details see the file of Tse3 element).
Branch 2 (non-chromodomain retroviruses and LTR retrotransposons)
The second Ty3/Gypsy branch encompasses the remaining lineages of non-chromoviral Ty3/Gypsy LTR retrotransposons and plant and animal retroviruses.
Athila/Tat
Within Branch 2, Athila/Tat is an ancient lineage that may be divided in two phylogenetically related lineages "Athila" and "Tat" (Wright and Voytas 1998; 2002). These two differ in that the elements belonging to the Athila clade usually code for an additional putative env gene and are considered as potential retroviruses, while Tat-like elements have the status of giant LTR retrotransposons.
Athila-like
Athila-like elements are 8.5-12 Kb in size, are flanked by LTRs of 1.5-2.5 Kb and have an internal region displaying a PBS, two ORFs for the gag and pol genes and, also a third one for a putative env gene already under study, and two PPTs adjacent to the 3´LTR.
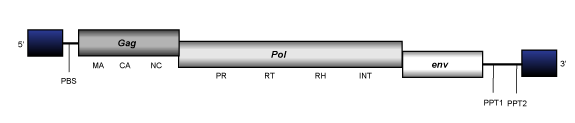
Tat-like
Tat-like elements are giant LTR retrotransposons with 10-21 Kb in size. They are flanked by LTRs of 0.5-1 Kb and have an internal region containing a PBS, gag and pol genes and a non-coding region of variable size, and a PPT adjacent to the 3´LTR. Some Tat retrotransposons present in the untranslated region an antisense ORFs of which little is known, and also, (Havecker, Gao, and Voytas 2004).
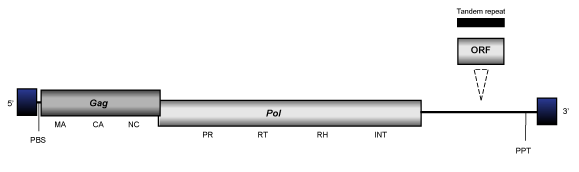
CsRN1
CsRN1 clade (Bae et al. 2001) is an ancient branch 2 lineage of bilateria-Ty3/Gypsy elements present in the genomes of certain protostome organisms. At the genomic structure level, the full length consensus for these elements presents an internal region of 5-6 Kb in size, flanked by LTRs of 0.1-0.5 Kb and characterized by the usual presence of a PBS, gag and pol genes, and a PPT adjacent to the 3´LTR.
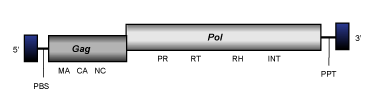
412/mdg1
412/mdg1 clade (Malik and Eickbush 1999) is a branch 2 lineage of Ty3/Gypsy LTR retrotransposons closely related to errantiviruses and described in arthropods. At the genomic structure level, 412/mdg1 elements present an internal region of 6-7.5 Kb in size, flanked by LTRs of 0.3-0.4 Kb and characterized by the usual presence of a PBS, gag and pol genes, and a PPT adjacent to the 3´LTR.
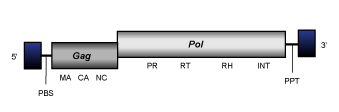
Errantiviruses
Within Branch 2, errantiviruses also known Gypsy-like elements, constitute the genus Errantiviridae (Boeke et al. 1999; Pringle 1998; 1999; Hull 1999), one of the most widely studied lineages of Ty3/Gypsy elements in arthropods. Errantiviruses collect both retroviruses and LTR retrotransposons. At the genomic structure level errantiviruses display an internal region of 7.8 Kb in size, flanked by LTRs of 0.3-0.6 Kb and characterized by the presence of a PBS, three ORFs for the gag, pol, and env genes typically observed in retroviruses, and also, a PPT adjacent to the 3´LTR. Many errantiviruses have not the third ORF and are, thus considered to be retrotransposons.
17.6
17.6 elements constitute a clade of LTR retrotransposons normally of 7-8 Kb and LTRs of 400 nucleotides (nts) in size. 17.6 retroviruses present an internal region displaying both a PBS, normally complementary to a tRNASerine, and a PPT and three ORFs for the typical gag, pol and env proteins coded by Errantiviridae elements.
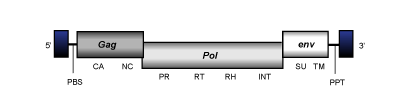
Gypsy
Gypsy is a clade collecting both retroviruses and LTR retrotransposons. They usually show genomes of 6-7 Kb and LTRs of 200-400 nts in size. Their internal region usually is arranged in two or three ORFs (depending if they include or not an env gene) flanked by a PBS normally complementary to a tRNALys, and a PPT downstream and upstream to the 5´and 3´LTRs, respectively.
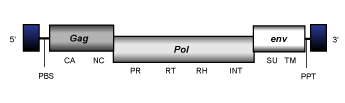
Osvaldo
Osvaldo clade (Malik and Eickbush 1999) is a branch 2 lineage that includes certain LTR retrotransposons described in arthropods and at least a fly-retrovirus called Osvaldo (Pantazidis et al. 1999). The genomic structure of Osvaldo-like elements is 7.8 Kb in size. It is flanked by LTRs of 0.3-0.6 Kb and exhibits a PBS, the usual gag and pol genes of LTR retrotransposons, except in the case of the Osvaldo species which displays an additional third ORF coding for an env polyprotein. And also, a PPT adjacent to the 3´LTR.
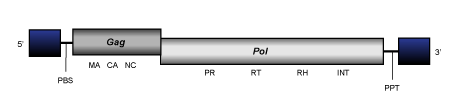
Gmr1
Gmr1 is an atypical clade of Ty3/Gypsy LTR retrotransposons to date only described in deuterostomes (Butler et al. 2001;Goodwin and Poulter 2002). Both phylogenetic and similarity analyses reveal that Gmr1 elements are apparently related to Osvaldo clade, a lineage of protostomes, but no Gmr1 representative has been described in protostomes. Interestingly, Gmr1 elements are atypical in the fact that although they are Ty3/Gypsy elements they functionally and evolutionarily preserve an ORFs organization identical to that of Ty1/Copia LTR retroelements (integrase after protease).
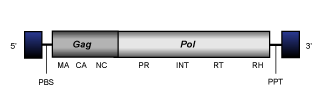
Cer2-3
Cer2-3 clade is a small branch 2 lineage of Ty3/Gypsy LTR retrotransposons present in the genome of Caenorhabditis elegans (Bowen and McDonald 1999). Cer2-3 elements present an internal region of 8.2-9.3 Kb in size, flanked by LTRs of 0.4-0.6 Kb and characterized by the presence of a PBS, the gag and pol genes typically observed in LTR retrotransposons, and a PPT adjacent to the 3´LTR.
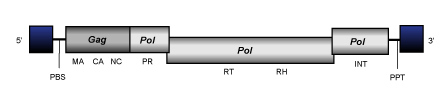
Cer1
Cer1 clade is a branch 2 lineage of Ty3/Gypsy LTR retroelements present in C. elegans and constituted by only one representative that gives the name to the clade (Britten 1995; Bowen and McDonald 1999). At the genomic structure level, Cer1 presents an internal region of 8.9 Kb in size, which is flanked by LTRs of approximately 0,5 Kb and characterized by the presence of a PBS, a single ORF containing the gag and pol genes and a PPT adjacent to the 3´LTR (for more details see the file of Cer1 element).
Mag
Mag (Malik and Eickbush 1999) is the most polyphyletic and widespread lineage of branch 2 bilateria-Ty3/Gypsy retrotransposons (Llorens et al. 2009), and has been described in the genomes of certain protostomes and echinoderms (Michaille et al. 1990; Springer, Davidson and Britten 1991; Bowen and McDonald 1999). However, recent studies reveal the presence of pseudogenized Mag copies in flies (Tubio, Naveira and Costas 2005), and presence of functional copies in several vertebrate genomes (Volff et al. 2001). At the genomic structure level, Mag elements show an internal region of 4-5 Kb in size, flanked by LTRs of 0.2 Kb and exhibiting a PBS, gag and pol genes, and a PPT adjacent to the 3´LTR. Phylogenetic analyses performed based on both gag and pol polyproteins (Llorens et al. 2009) reveal three major clades among Mag elements:
A-clade
This clade includes both deuterostome and protostome LTR retrotransposons, all of them usually characterized by displaying small genomes (around 5 Kb) and LTRs (beetwen 130-190 nts) with an internal region that may be organized in one, two or three ORFs, as well as the typical PBS and PPT upstream and downstream to the 5´and 3´LTRs, respectively.
B-clade
Based on current evidence, this clade includes only protostome LTR retrotransposons, all of them showing the typical size of Mag-like retrotransposons (around 5 kb) and small LTRs (over 165-200 nts). The internal region can display one, two or three ORFs encoding the typical gag and pol products and features such as a PBS, which may complementary to a tRNASer or a a tRNAMet, and the PPT.
C-clade
To date, C-clade is represented by SURL (Springer, Davidson and Britten 1991), a deuterostome LTR retrotransposon of 5.3 Kb in size organized in a gag-pol internal region flanked by LTRs of 0.2 Kb and displaying both PBS (complementary to a tRNASer) and PPTs downstream and upstream of the 5´ and 3´LTRs, respectively (for more details see the file of SURL element).
Tor1
Tor1 (Ty3/Gypsy Oikopleura retrotransposons Type 1) is a single-element lineage with at least two copies described in Oikopleura dioica (Volff et al. 2004). The elements belonging to Tor1 display the typical features of LTR retrotransposons and seem more similar to the Ty3/Gypsy lineages of protostomes than to those of deuterostomes. It includes the remaining Tor-like elements described in Oikopleura dioica, which differ each other not only in sequence similarity but also in molecular phenotypes such as variability in zinc finger arrays at the gag-nucleocapsid, presence of env, etc (for more details see the file of Tor1 element).
Tor2
Tor2 (Ty3/Gypsy Oikopleura retrotransposons Type 2) is another Ty3/Gypsy lineage of LTR retrotransposons described but more widely distributed than Tor1 in Oikopleura dioica (Volff et al. 2004). The distinct copies of Tor2 display between 60-90% of nucleotide identity and show the typical features of LTR retrotransposons. As other Tor-like lineages, Tor2 elements are more similar to diverse Ty3/Gypsy lineages of protostomes than to those described in deuterostomes (for more details see the file of Tor2 element).
Tor4
Tor4 (Ty3/Gypsy Oikopleura retrotransposons type 4) constitutes another lineage within the diversity of Ty3/Gypsy retrotransposons and retroviruses described in the genome Oikopleura dioica (Volff et al. 2004). Tor4 collects both LTR retrotransposons and potential retroviruses. We take Tor4a as a representative sequence (a LTR retrotransposon displaying the typical gag and pol ORFs) but other retroviral variants of this lineage are described in Volff et al. (2004) together with Tor3, which is another potential Ty3/Gypsy lineage of retroviruses not yet classified in this project (for more details see the file of Tor4a element).
Cigr-1
Cigr-1 is a heterogeneous Ty3/Gypsy LTR retrotransposon described in the genome of Ciona intestinalis (Simmen et al. 1998;1999; Simmen and Bird 2000) and constitutes an own Ty3/Gypsy single-element-lineage. Its phylogenetic signal varies, however, depending on the gag and pol polyproteins. While pol-based phylogenies suggest that Cigr-1 is tentatively but not conclusively similar to Errantiviruses (and others), both phylogenetic and comparative analyses based on gag clearly reveal that Cigr-1 is counterpart of Mag-like Ty3/gypsy elements (Llorens et al. 2008a; 2008b). In other words, Cigr-1 clade is probably a distant sibling of Mag or the chimeric result of an ancient recombination event (for more details see the file of Cigr-1 element).
Micropia/Mdg3
Micropia/mdg3 clade (Malik and Eickbush 1999) is a branch 2 lineage exclusively constituted by LTR retrotransposons, which at the genomic structure level exhibit an internal region of approximately 5 Kb in size, flanked by LTRs of 0.2-0.5 Kb and characterized by the presence of a PBS, the usual gag and pol genes, and a PPT adjacent to its 3´LTR.