Description
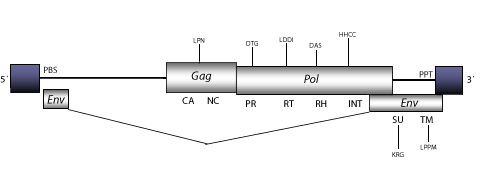
Zam is a retrovirus originally identified as a mutational insertion in the white locus of Drosophila melanogaster. This retrovirus is mobilized in the germinal lines (Leblanc et al. 1997) and several studies have also reported mobilization of Zam (and Idefix) in the line RevI of Drosophila melanogaster (Desset et al. 1999; Conte et al. 2000). Zam belongs to the genus Errantiviridae (Boeke et al. 1999; Pringle 1998; 1999; Hull 1999), a lineage of fly-retroviruses and LTR retrotransposons closely related to 412/mdg1 clade (Tubio, Naveira and Costas 2005). The genomic structure of Zam is 8.6 Kb in size including LTRs of 472 nt. The internal region displays a Primer Binding Site (PBS) complementary to a tRNASer, two Open Reading Frames (ORFs) for gag and pol genes, two ORFs that codify for a subgenomic env product, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Leblanc et al. 1997).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|