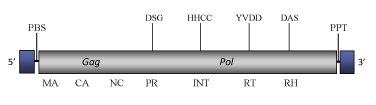
Element:Tdh2
DescriptionTdh2, a Ty1/Copia-like LTR retrotransposon characterized in the genome of the spoilage yeast Debaryomyces hansenii (Neuvéglise et al. 2002). This element belongs to Ty (Pseudovirus) clade one of the three original genera on which the in-progrees Pseudoviridae classification is constituted (Boeke et al 2005). The genome of Tdh2 is 5928 bp in size, including two 379-bp LTRs that bound an internal coding region containing a Primer Binding Site (PBS), complementary to a tRNAArg, a long Open Reading Frame (ORF) interrupted by one stop codon, and a Polypurine Tract (PPT) upstream to the 3’LTR (Neuvéglise et al. 2002). The ORF shows the typical domains associated to Ty1/Copia gag and pol genes. The gag-associated RNA-binding motif (CCHC) has not been identified in this element (Neuvéglise et al. 2002). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||