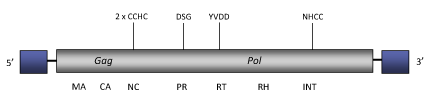
Element:Sinbad
DescriptionSinbad is a LTR retrotransposon characterized in Schistosoma mansoni (Copeland et al. 2005) and also present in S. haematobium (Copeland et al. 2006). About 50 copies of this element appear to reside in the S. mansoni genome. The term Sinbad term derives from the legendary mariner-explorer Sinbad protagonist of the classical Persian/Arabic tales "1001 Arabian Nights" (Sinbad traveled through near Eastern countries where schistosomiasis disease remains endemic even today). Sinbad gives the name and belongs to Sinbad clade (Copeland et al. 2005). The genome structure of Sinbad is about 6.3 Kb in size, includes LTRs of 386 nt long and encodes for a single ORF. Although the internal coding region is disrupted by several stop codons and frameshiftings, the distinct gag and pol domains typical of LTR retrotransposons are clearly evident in the sequence of Sinbad (Copeland et al. 2005). The figure below shows an idealized consensus structure. Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||