Bel/Pao
Introduction:
Bel/Pao LTR retroelements (Malik et al. 2000) constitute a family of LTR retrotransposons and retroviruses described to date only in metazoan genomes (Copeland et al. 2005). This family was originally characterized with the discovery of element sequences such as Pao (Xiong et al. 1993), Bel (Davis and Judd 1995), and Tas (Aeby et al. 1986) and the various Cer-like sequences described in Caenorhabditis elegans (Bowen and McDonald 1999). At present and according with the International Committee on the Taxonomy of Viruses (ICTV), the multiple distinct members of the Bel/Pao family are called semotiviruses (for a review, see Eickbush and Jamburuthugoda 2008).
Genomic structure:
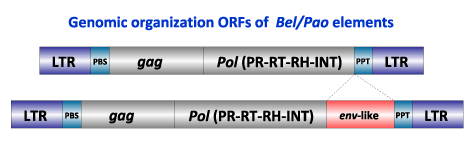
Similarly to the Ty3/Gypsy and the Retroviridae, Bel/Pao LTR retroelements normally show a gag-pol genome (GAG-PR-RT-RH-INT) plus env (in the case of retroviruses) of variable size (between 4 and 10 Kb), flanked by LTRs and structured (in its inserted genomic form) as follows:
- A 5' long terminal repeat (LTR) of 100-900 nt
- A non-coding region of variable size that corresponds to the first portion of the retrotranscribed genome.
- A Primer Binding Site (PBS) of 18 nt, complementary to a specific zone of the 3' end of a tRNA, normally provided by the host cell in order to start the retrotranscription.
- Open Reading Frames (ORFs) for gag, pol, (and env in retroviruses) genes.
- A small region of ~10 A/G "Polypurine Tract" (PPT), responsible for starting the synthesis of the proviral DNA strand.
- A 3' long terminal repeat (LTR) of 100-900 nt
Evolutionary history and in-progress taxonomy:
The Bel/Pao family can be divided into five lineages called Pao, Sinbad, Bel, Tas and Suzu (Copeland et al. 2005). Phylogenetic approaches based on pol (Llorens et al. 2009) show that these five can be divided into three branches namely as 1, 2 and 3. The first includes the two clades Tas and Bel, the second collects Pao and Sinbad clades, and the last consists of a single clade called Suzu.
| Branch | Host Phyla | Genus | Clade | Env |
|---|---|---|---|---|
| Branch 1 | Metazoa (Nematoda and Cnidaria) | Semotivirus | Tas | |
| Branch 1 | Metazoa (Insecta) | Semotivirus | Bel | |
| Branch 2 | Metazoa (Insecta and Vertebrata) | Semotivirus | Pao | |
| Branch 2 | Metazoa (Invertebrata) | Semotivirus | Sinbad | |
| Branch 3 | Metazoa (Echinodermata and Vertebrata) | Semotivirus | Suzu |
Branch 1
Branch 1 encompasses two clades of LTR retrotransposons and potential retroviruses called Tas and Bel found in the genomes of cnidarians and protostomes.
Tas
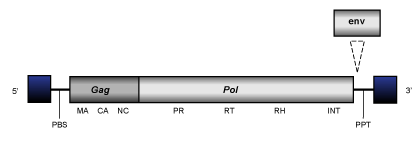
Tas clade (Copeland et al. 2005) is constituted by diverse Bel/Pao-like LTR retrotransposons and retroviruses described in nematodes and cnidarians (Bowen and McDonald 1999; Felder et al. 1994; Llorens et al. 2009). Their genome is about of 5.4-10 Kb in size including, in almost (but not all) retroelements, LTRs of 0.23-0.81 Kb long. The internal coding region is respectively flanked by a Primer Binding Site (PBS) and a Polypurine Tract (PPT) and, although is usually disrupted by several stop codons and frameshiftings, it displays the distinct gag and pol domains typical of Bel/Pao LTR retrotransposons. In some elements of this clade, it has also been identified an env-like gene located at their C-terminal translated regions (Felder et al. 1994; Bowen and McDonald 1999). (The figure below shows an idealized consensus of Tas elements genome organization).
Bel
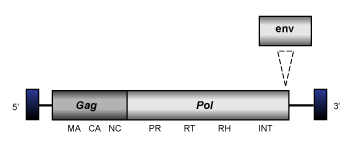
Bel (Copeland et al. 2005) is a lineage of insect LTR retrotransposons which usually are 4.6-9.0 Kb in size and show an internal region flanked by LTRs of 0.2-0.4 kb long. The members of this clade may encode for one, two or three ORFs displaying the typical gag-pol domains. Interestingly, our analyses reveal that the env coded by Roo (the Bel-like element represented in the figure below) is more similar to those coded by Ty3/Gypsy errantiviruses than to those coded by other Bel/Pao potential retroviruses. This suggests that several Bel/Pao elements might have evolved from the chimeric recombination between a Ty3/Gypsy retrovirus and a Bel/Pao retrotransposon.
Branch 2
Branch 2 includes the Pao and Sinbad clades of LTR retrotransposons described in protostomes and deuterostomes.
Pao
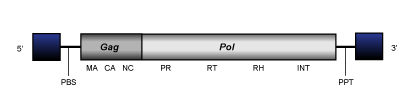
Pao is a lineage of LTR retrotransposons (Copeland et al. 2005) widely distributed in the genomes of several protostomes and deuterostomes. Pao elements usually have a size of 4.7-8.3 Kb (including LTRs of 0.3-1.3 Kb), their internal region is characterized by the usual presence of a PBS, gag-pol coding region and PPT.
Sinbad
Sinbad clade (Copeland et al. 2005) collects several LTR retrotransposons described in both protostome (Platyhelminthes) and deuterostome (Hemichordates) worms. Sinbad elements normally display genomes of 4.2-7.4 Kb in size, including flanking LTRs of 0.08-0.86 Kb and are characterized by an internal gag-pol region plus flanking PBS and PPT as is typical in Bel/Pao LTR retrotransposons.
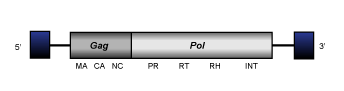
Branch 3
Branch 3 consists of a single clade called Suzu, which includes LTR retrotransposons that have so far been described exclusively in vertebrates.
Suzu
Suzu clade (Copeland et al. 2005) is constituted by diverse deuterostome LTR retrotrasposons (described in echinoderms and vertebrates). Suzu-like elements usually are about 7-7.6 Kb in size including LTRs of 0.4-0.7 kb long and code for the typical gag and pol protein domains.

