Element:DrMV
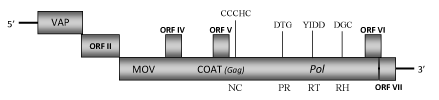
DescriptionDracaena mottle virus (DrMV) is the tentative name associated to a plant pararetrovirus infecting the indoor ornamental plant "Fortune Bamboo" (Dracaena sanderiana) in which provokes typical symptoms such as mottling and chlorotic patches on leaves (Su et al. 2007). DrMV is a member of genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 is located within Class 2 of the Caulimoviridae family. DrMV has bacilliform-shaped particles of approximately 125–130 nm x 30 nm containing a dsDNA genome which size and organization are similar (but not identical) to those of other badnaviruses (Su et al. 2007). Unlike most badnaviruses that contain the typical three open reading frames (ORFI, II, III), DrMV also encodes for other additional ORFs (ORF IV, V, VI and VII) (Su et al. 2007). The product of ORFI of 17.6 kDa shows sequence similarities to ORFI-virion associated proteins (VAPs) usually present in the genome of Caulimoviridae species; ORF II potentially encodes a protein of 131 amino acids (aa) which function has not been yet demonstrated even if it has been described a putative capacity to bind nucleic acids for ORF II products of Cacao swollen shoot virus (CSSV) and Rice tungro bacilliform virus (RTBV) (Jacquot et al. 1996; Jacquot et al. 1997). ORF III contains a polyprotein with the typical motifs highly conserved amongst badnavirus proteins [the MOV, the COAT zinc finger-like RNA-binding motif (C-X-C-X2-C-X4-H-X4-C) and the second large cysteine-rich region, the PR, the RT and the RH] (Su et al. 2007). The large region rich in zinc finger (CCHC) array duplications (only observed in Badna- and Tungroviruses), presents in the COAT gag-like region of this element resembles to those identified in the nucleocapsid domains of LTR retroelements (Llorens et al. 2009). Other small addicional ORFs displayed by DrMV genome potentially encode for protein with a molecular weight of 11.9 kDa (ORF IV), 11.3 kDa (ORF V), 16.1 kDa (ORF VI) and 11.0 kDa (ORF VII). Although a fourth ORF is also present in the genome of TaBV, CSSV and Citrus yellow mosaic virus (CMBV) badnaviruses, only the gene product of CMBV-ORF IV shows a significant sequence homology with that of DrMV (66.7%)(Su et al. 2007). Like DrMV, more than one additional ORF has been also found in the genome of other badnaviruses with which DrMV-additional ORFs have shown some similarities. DrMV-ORF V share similarities at both the nucleotide and amino acid levels (38% and 40%, respectively) with CMBV-ORF V, as well as similarities among DrMV-ORF VI and CSSV-ORF Y and CMBV-ORF VI were also found (Su et al. 2007). Our sequence analyses reveal that the presence of a stop codon at the C-terminus of ORF III generates the small ORF VII that has not been found in all the badnaviruses sequenced so far. However, these protein products do not show any known function, so it remains unclear if they are a real viral gene products (Su et al. 2007). The presence of pararetroviral sequences integrated in the host genome has been frequently reported (Bousalem et al. 2009; Kenyon et al. 2008; Gayral et al. 2008; Geering et al. 2005; Kunii et al. 2004; Yang et al. 2003; Harper et al. 2002; Lockhart et al. 2000; Harper et al. 1999; Richert-Pöggeler and Shepherd 1997). Multiple copies of DrMV sequences (corresponding to the region from nt 775 to nt 6040) have been also detected in the genomic DNA of asymptomatic D. sanderiana plants. These results suggest that the DrMV sequences might be integrated into the host genome but remain to confirm if a complete DrMV sequence is integrated in the host genome and if this integrated sequence is capable of giving rise to a functional episomal genome (Su et al. 2007). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||